Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
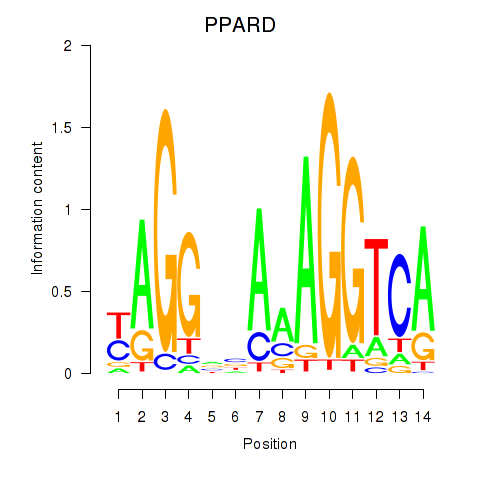
Results for PPARD
Z-value: 0.43
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARD | hg19_v2_chr6_+_35310391_35310410 | 0.97 | 2.6e-02 | Click! |
Activity profile of PPARD motif
Sorted Z-values of PPARD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_76030311 | 0.34 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr19_-_49339080 | 0.21 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_+_145727681 | 0.21 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr17_+_7123207 | 0.19 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr15_+_82555125 | 0.17 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr7_+_7222157 | 0.16 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr17_-_71223839 | 0.16 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr3_-_53878644 | 0.16 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr11_+_64073699 | 0.16 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr3_-_50605077 | 0.15 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr19_+_10196981 | 0.15 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_+_111169565 | 0.14 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr11_-_45939565 | 0.13 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr12_+_121163602 | 0.12 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr7_+_75911902 | 0.12 |
ENST00000413003.1
|
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr9_-_33402506 | 0.12 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr16_+_81272287 | 0.12 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr12_-_109219937 | 0.12 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr1_+_43855545 | 0.12 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr19_-_10687907 | 0.11 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr2_+_159651821 | 0.10 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr11_+_111169976 | 0.10 |
ENST00000398035.2
|
COLCA2
|
colorectal cancer associated 2 |
| chr17_+_9745786 | 0.10 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr3_-_49314640 | 0.10 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr5_-_143550241 | 0.10 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr12_-_109797249 | 0.09 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr11_-_45939374 | 0.09 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr12_+_121163538 | 0.08 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr11_-_116968987 | 0.08 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr6_+_31637944 | 0.07 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr3_-_49203744 | 0.07 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr16_-_75284758 | 0.07 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_-_61659006 | 0.06 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr6_-_33548006 | 0.06 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr5_-_176981417 | 0.06 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr6_-_33547975 | 0.06 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr17_+_48610074 | 0.05 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr19_+_10197463 | 0.05 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr10_-_103815874 | 0.05 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr2_-_198364552 | 0.05 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr19_+_29493486 | 0.05 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr2_+_198365122 | 0.05 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr11_-_61658853 | 0.05 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr8_-_135652051 | 0.05 |
ENST00000522257.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr11_-_47470703 | 0.05 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr17_+_7123125 | 0.04 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr9_+_131901661 | 0.04 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr15_+_91449971 | 0.04 |
ENST00000557865.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr19_+_49867181 | 0.04 |
ENST00000597546.1
|
DKKL1
|
dickkopf-like 1 |
| chr1_+_202317815 | 0.04 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr13_+_113760098 | 0.04 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr5_+_14664762 | 0.04 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr16_-_28303360 | 0.04 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr2_+_95940186 | 0.04 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr15_+_75494214 | 0.03 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr19_-_51869592 | 0.03 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr1_-_153917700 | 0.03 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr16_-_4401284 | 0.03 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr20_+_39969519 | 0.03 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chr1_+_43855560 | 0.03 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr14_-_51561784 | 0.03 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr11_-_119599794 | 0.03 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr17_-_7123021 | 0.03 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr20_+_32077880 | 0.03 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr1_-_11863171 | 0.03 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr4_+_140586922 | 0.02 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr10_-_94050820 | 0.02 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr16_-_74808710 | 0.02 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr2_+_178257372 | 0.02 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chrX_-_108868390 | 0.02 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr6_-_135424186 | 0.02 |
ENST00000529882.1
|
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr21_-_27107283 | 0.02 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chrX_+_2746818 | 0.02 |
ENST00000398806.3
|
GYG2
|
glycogenin 2 |
| chr17_+_37894179 | 0.02 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr2_+_39117010 | 0.02 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr2_-_159313214 | 0.02 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr20_+_46130601 | 0.02 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr11_-_47470591 | 0.02 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr2_-_207024134 | 0.02 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr20_-_44485835 | 0.02 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr1_+_244998602 | 0.01 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr19_-_10687948 | 0.01 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_11546153 | 0.01 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr19_-_10687983 | 0.01 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr12_-_56727487 | 0.01 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_+_120125245 | 0.01 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr19_+_29493456 | 0.01 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr15_-_43865633 | 0.01 |
ENST00000437065.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr6_+_30295036 | 0.01 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr12_+_60058458 | 0.01 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr19_+_11546440 | 0.01 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chrX_+_2746850 | 0.01 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr19_+_11546093 | 0.01 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr14_+_74551650 | 0.01 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr1_-_156675564 | 0.01 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr11_+_123325106 | 0.00 |
ENST00000525757.1
|
LINC01059
|
long intergenic non-protein coding RNA 1059 |
| chr19_-_3062465 | 0.00 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr16_-_75285380 | 0.00 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr5_-_16509101 | 0.00 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr11_+_17281900 | 0.00 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |