Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
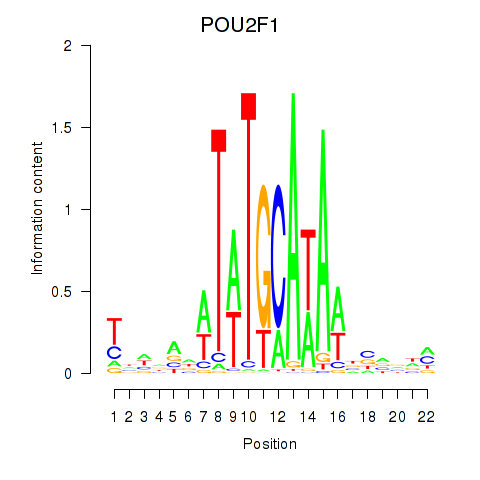
Results for POU2F1
Z-value: 0.87
Transcription factors associated with POU2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F1
|
ENSG00000143190.17 | POU class 2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F1 | hg19_v2_chr1_+_167190066_167190156 | -0.75 | 2.5e-01 | Click! |
Activity profile of POU2F1 motif
Sorted Z-values of POU2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_13621126 | 0.50 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr6_+_27782788 | 0.35 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr3_+_151591422 | 0.34 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr8_+_86121448 | 0.34 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr6_-_26033796 | 0.29 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr16_-_18462221 | 0.26 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr1_+_104159999 | 0.26 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr3_-_121379739 | 0.21 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_+_90458201 | 0.20 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr12_+_11187087 | 0.20 |
ENST00000601123.1
|
AC018630.1
|
Uncharacterized protein |
| chr15_+_80364901 | 0.19 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr6_+_122720681 | 0.18 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr2_+_173292059 | 0.18 |
ENST00000412899.1
ENST00000409532.1 |
ITGA6
|
integrin, alpha 6 |
| chr6_-_26216872 | 0.18 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr22_-_37571089 | 0.17 |
ENST00000453962.1
ENST00000429622.1 ENST00000445595.1 |
IL2RB
|
interleukin 2 receptor, beta |
| chr1_+_62439037 | 0.17 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr8_-_101730061 | 0.17 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_+_75230853 | 0.17 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr3_+_32737027 | 0.16 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr2_-_74007095 | 0.15 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr5_+_112227311 | 0.15 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr12_-_106480587 | 0.15 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr3_-_145940126 | 0.15 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr22_+_23029188 | 0.15 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr3_-_167191814 | 0.14 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr12_-_111358372 | 0.14 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr1_-_17231271 | 0.14 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr16_+_14980632 | 0.14 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr2_+_62900986 | 0.14 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr2_+_169659121 | 0.14 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr11_+_94300474 | 0.13 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr1_-_213020991 | 0.13 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr2_-_4703793 | 0.13 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr12_+_40618873 | 0.13 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr3_+_149530836 | 0.12 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chrY_+_14958970 | 0.12 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr6_+_135502408 | 0.12 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr9_-_116065551 | 0.12 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr11_+_85359062 | 0.12 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr12_+_16500599 | 0.12 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr3_+_118892411 | 0.12 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr16_+_33629600 | 0.12 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr8_+_30244580 | 0.11 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr15_-_50838888 | 0.11 |
ENST00000532404.1
|
USP50
|
ubiquitin specific peptidase 50 |
| chr11_-_107729887 | 0.11 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_-_148939835 | 0.11 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr7_-_103848405 | 0.11 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr2_+_169658928 | 0.11 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_+_197464046 | 0.11 |
ENST00000428738.1
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr7_-_99149715 | 0.10 |
ENST00000449309.1
|
FAM200A
|
family with sequence similarity 200, member A |
| chr18_+_61144160 | 0.10 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr6_-_26124138 | 0.10 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr2_-_74007193 | 0.10 |
ENST00000377706.4
ENST00000443070.1 ENST00000272444.3 |
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr2_-_61108449 | 0.10 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr13_+_108870714 | 0.10 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr8_+_18067602 | 0.10 |
ENST00000307719.4
ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr19_+_14693888 | 0.10 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr11_-_125366018 | 0.10 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr7_+_64838786 | 0.10 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr2_+_109237717 | 0.10 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_48594248 | 0.10 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr8_-_117768023 | 0.10 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr16_+_81070792 | 0.09 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr9_+_124088860 | 0.09 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr3_+_157154578 | 0.09 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_107729504 | 0.09 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_-_107729287 | 0.09 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr21_-_44582378 | 0.09 |
ENST00000433840.1
|
AP001631.10
|
AP001631.10 |
| chr3_-_155524049 | 0.09 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr5_+_61874562 | 0.09 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr1_+_100436065 | 0.09 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr12_-_58329819 | 0.09 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr12_-_91546926 | 0.09 |
ENST00000550758.1
|
DCN
|
decorin |
| chr2_-_236684438 | 0.09 |
ENST00000409661.1
|
AC064874.1
|
Uncharacterized protein |
| chr3_+_118892362 | 0.09 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr3_+_136676851 | 0.09 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr5_-_1551802 | 0.09 |
ENST00000503113.1
|
CTD-2245E15.3
|
CTD-2245E15.3 |
| chr20_+_36888551 | 0.08 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr3_-_58200398 | 0.08 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr5_-_176326333 | 0.08 |
ENST00000292432.5
|
HK3
|
hexokinase 3 (white cell) |
| chr13_+_24883703 | 0.08 |
ENST00000332018.4
|
C1QTNF9
|
C1q and tumor necrosis factor related protein 9 |
| chr14_+_89290965 | 0.08 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr14_-_92247032 | 0.08 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr16_+_73420942 | 0.08 |
ENST00000554640.1
ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1
|
RP11-140I24.1 |
| chr20_+_39657454 | 0.08 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr16_-_46782221 | 0.08 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr2_-_145275109 | 0.08 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_-_19988382 | 0.07 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chrX_+_119384607 | 0.07 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr8_-_117886612 | 0.07 |
ENST00000520992.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr6_+_135502501 | 0.07 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_110906027 | 0.07 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr16_-_31228680 | 0.07 |
ENST00000302964.3
|
PYDC1
|
PYD (pyrin domain) containing 1 |
| chr7_-_14026063 | 0.07 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr2_+_183582774 | 0.07 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr19_-_14682838 | 0.07 |
ENST00000215565.2
|
NDUFB7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa |
| chr4_+_74301880 | 0.07 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr10_+_79793518 | 0.06 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr14_-_65346555 | 0.06 |
ENST00000542895.1
ENST00000556626.1 |
SPTB
|
spectrin, beta, erythrocytic |
| chrX_-_130423240 | 0.06 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr16_+_16434185 | 0.06 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr12_+_105380073 | 0.06 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr20_+_5731027 | 0.06 |
ENST00000378979.4
ENST00000303142.6 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr12_+_40618764 | 0.06 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr4_-_144940477 | 0.06 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr7_+_64838712 | 0.06 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr20_+_61427797 | 0.06 |
ENST00000370487.3
|
MRGBP
|
MRG/MORF4L binding protein |
| chr15_-_63448973 | 0.06 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr12_+_72056773 | 0.06 |
ENST00000308086.2
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr7_+_7606497 | 0.06 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chrX_-_130423386 | 0.06 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chrX_-_130423200 | 0.06 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr13_+_111855414 | 0.05 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_+_67588391 | 0.05 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr2_-_113191096 | 0.05 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr12_-_58329888 | 0.05 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr17_+_45728427 | 0.05 |
ENST00000540627.1
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr21_+_17553910 | 0.05 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr10_-_115904361 | 0.05 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr13_-_20110902 | 0.05 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr11_+_3011093 | 0.05 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr15_-_55541227 | 0.05 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_+_126626498 | 0.05 |
ENST00000503335.2
ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10
|
multiple EGF-like-domains 10 |
| chr2_+_110551851 | 0.05 |
ENST00000272454.6
ENST00000430736.1 ENST00000016946.3 ENST00000441344.1 |
RGPD5
|
RANBP2-like and GRIP domain containing 5 |
| chr2_-_145275211 | 0.05 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_128457446 | 0.05 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr22_-_29949680 | 0.05 |
ENST00000397873.2
ENST00000490103.1 |
THOC5
|
THO complex 5 |
| chr17_-_41910505 | 0.05 |
ENST00000398389.4
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr22_-_29949657 | 0.05 |
ENST00000428374.1
|
THOC5
|
THO complex 5 |
| chr11_-_88799113 | 0.05 |
ENST00000393294.3
|
GRM5
|
glutamate receptor, metabotropic 5 |
| chr3_+_149531607 | 0.05 |
ENST00000468648.1
ENST00000459632.1 |
RNF13
|
ring finger protein 13 |
| chr11_+_120107344 | 0.04 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chrX_-_99665262 | 0.04 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr2_+_71205742 | 0.04 |
ENST00000360589.3
|
ANKRD53
|
ankyrin repeat domain 53 |
| chr15_+_43425672 | 0.04 |
ENST00000260403.2
|
TMEM62
|
transmembrane protein 62 |
| chr2_-_44550441 | 0.04 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr10_+_13628933 | 0.04 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr3_+_182983126 | 0.04 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr4_-_120550146 | 0.04 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr10_+_126630692 | 0.04 |
ENST00000359653.4
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1 |
| chr10_-_75118471 | 0.04 |
ENST00000340329.3
|
TTC18
|
tetratricopeptide repeat domain 18 |
| chr1_-_146082633 | 0.04 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chrX_-_135962876 | 0.04 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr7_-_14026123 | 0.04 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr3_+_196466710 | 0.04 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr18_+_44812072 | 0.04 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr10_+_26932068 | 0.04 |
ENST00000544033.1
ENST00000454991.1 |
LINC00202-2
RP13-16H11.7
|
long intergenic non-protein coding RNA 202-2 RP13-16H11.7 |
| chrX_+_134478706 | 0.04 |
ENST00000370761.3
ENST00000339249.4 ENST00000370760.3 |
ZNF449
|
zinc finger protein 449 |
| chr16_+_14294187 | 0.04 |
ENST00000573051.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr9_+_112887772 | 0.04 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chrX_+_57313113 | 0.04 |
ENST00000374900.4
|
FAAH2
|
fatty acid amide hydrolase 2 |
| chr12_-_50616382 | 0.03 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr10_+_91589261 | 0.03 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr6_+_116832789 | 0.03 |
ENST00000368599.3
|
FAM26E
|
family with sequence similarity 26, member E |
| chr9_-_123812542 | 0.03 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr9_+_116638630 | 0.03 |
ENST00000452710.1
ENST00000374124.4 |
ZNF618
|
zinc finger protein 618 |
| chr12_+_27849378 | 0.03 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr22_-_16449805 | 0.03 |
ENST00000252835.4
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1 |
| chr16_-_90142338 | 0.03 |
ENST00000407825.1
ENST00000449207.2 |
PRDM7
|
PR domain containing 7 |
| chr6_+_106988986 | 0.03 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr10_-_52008313 | 0.03 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr20_+_9146969 | 0.03 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr2_-_152684977 | 0.03 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr3_+_187896331 | 0.03 |
ENST00000392468.2
|
AC022498.1
|
Uncharacterized protein |
| chr10_+_91589309 | 0.03 |
ENST00000448490.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr3_+_101546827 | 0.03 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr18_-_61311485 | 0.03 |
ENST00000436264.1
ENST00000356424.6 ENST00000341074.5 |
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr1_-_201123546 | 0.03 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr4_+_40194609 | 0.03 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr20_+_61904556 | 0.03 |
ENST00000522403.2
ENST00000550188.1 |
ARFGAP1
|
ADP-ribosylation factor GTPase activating protein 1 |
| chr1_-_6614565 | 0.03 |
ENST00000377705.5
|
NOL9
|
nucleolar protein 9 |
| chr4_+_40194570 | 0.03 |
ENST00000507851.1
|
RHOH
|
ras homolog family member H |
| chr14_+_19377522 | 0.03 |
ENST00000550708.1
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12 |
| chr2_-_197226875 | 0.02 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_-_7818520 | 0.02 |
ENST00000329434.2
|
OR5P2
|
olfactory receptor, family 5, subfamily P, member 2 |
| chr12_+_27623565 | 0.02 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr4_-_152147579 | 0.02 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr17_+_75181292 | 0.02 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr10_+_13628921 | 0.02 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr5_-_35230649 | 0.02 |
ENST00000382002.5
|
PRLR
|
prolactin receptor |
| chrX_+_114827851 | 0.02 |
ENST00000539310.1
|
PLS3
|
plastin 3 |
| chr1_-_101360331 | 0.02 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr6_-_46922659 | 0.02 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chrX_-_11369656 | 0.02 |
ENST00000413512.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr2_-_111334678 | 0.02 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr7_+_90338712 | 0.02 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr9_-_85882145 | 0.02 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr12_-_95510743 | 0.02 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr18_+_616711 | 0.02 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr4_-_163085107 | 0.02 |
ENST00000379164.4
|
FSTL5
|
follistatin-like 5 |
| chr1_+_104293028 | 0.02 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr2_+_219524379 | 0.02 |
ENST00000443791.1
ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr16_-_18908196 | 0.02 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_+_161736072 | 0.02 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr19_+_39903185 | 0.02 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr1_-_157567868 | 0.02 |
ENST00000271532.1
|
FCRL4
|
Fc receptor-like 4 |
| chr3_+_136676707 | 0.02 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr2_+_102615416 | 0.02 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr3_-_111314230 | 0.02 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr6_-_135424186 | 0.01 |
ENST00000529882.1
|
HBS1L
|
HBS1-like (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.2 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.2 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.2 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.2 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |