Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
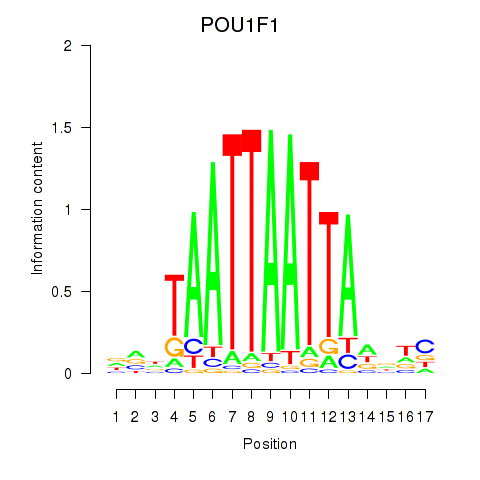
Results for POU1F1
Z-value: 0.76
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325728_87325739 | 0.60 | 4.0e-01 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13141010 | 0.97 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr12_-_25150373 | 0.83 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr19_-_58864848 | 0.70 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr12_+_78359999 | 0.62 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr8_-_145754428 | 0.50 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr6_+_26104104 | 0.46 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr6_-_26235206 | 0.44 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr19_+_52873166 | 0.41 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr20_-_29896388 | 0.40 |
ENST00000400549.1
|
DEFB116
|
defensin, beta 116 |
| chr19_+_50016411 | 0.38 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr9_-_95055923 | 0.37 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr4_-_120243545 | 0.35 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr19_+_50016610 | 0.34 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr8_-_124741451 | 0.32 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr17_+_1944790 | 0.31 |
ENST00000575162.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chr5_+_140762268 | 0.30 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr16_-_28634874 | 0.29 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr5_+_129083772 | 0.26 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr2_+_232575168 | 0.25 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr7_-_111032971 | 0.25 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_34175398 | 0.24 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr2_+_161993465 | 0.24 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr18_-_59274139 | 0.24 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr17_+_46189311 | 0.23 |
ENST00000582481.1
|
SNX11
|
sorting nexin 11 |
| chr17_-_72772462 | 0.22 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_+_35222629 | 0.22 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_+_47610315 | 0.22 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr17_+_79849872 | 0.21 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr11_+_59705928 | 0.21 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr22_+_23487513 | 0.20 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr6_+_26087646 | 0.19 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr3_+_52245458 | 0.19 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr6_+_114178512 | 0.18 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr12_-_52761262 | 0.18 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr6_+_26087509 | 0.18 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr12_-_11150474 | 0.18 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr19_+_36139125 | 0.18 |
ENST00000246554.3
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr2_-_166930131 | 0.17 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr16_-_28937027 | 0.17 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr10_+_18549645 | 0.17 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_+_15489603 | 0.17 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr7_-_22862448 | 0.17 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr12_+_113796347 | 0.16 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr6_-_152639479 | 0.16 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr12_+_38710555 | 0.16 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr17_+_78518617 | 0.16 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr9_-_33473882 | 0.16 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr11_-_559377 | 0.16 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr21_-_27423339 | 0.15 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr12_-_86650077 | 0.15 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_+_75639372 | 0.15 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr16_-_66584059 | 0.15 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr14_+_61449076 | 0.15 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr5_+_59783941 | 0.15 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr11_-_111649074 | 0.15 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr6_+_26440700 | 0.15 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr17_-_57158523 | 0.15 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr1_+_151253991 | 0.14 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr17_-_4938712 | 0.14 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr12_-_67197760 | 0.14 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr3_+_38537960 | 0.14 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr15_+_75970672 | 0.14 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr11_+_5710919 | 0.14 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr1_+_81106951 | 0.14 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr7_+_13141097 | 0.14 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr2_-_190044480 | 0.14 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr1_+_117963209 | 0.14 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr18_-_64271363 | 0.14 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr6_+_4087664 | 0.14 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr4_+_96012585 | 0.14 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_-_220264703 | 0.13 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr1_+_221051699 | 0.13 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr15_+_67418047 | 0.13 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr17_+_7155819 | 0.13 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr2_-_236684438 | 0.13 |
ENST00000409661.1
|
AC064874.1
|
Uncharacterized protein |
| chr7_-_84122033 | 0.12 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_-_215744 | 0.12 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr10_+_5135981 | 0.12 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr1_+_149754227 | 0.12 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr2_+_58655461 | 0.12 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr20_+_5986756 | 0.12 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr10_-_28623368 | 0.12 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr7_-_112758589 | 0.12 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr15_-_64385981 | 0.12 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr19_+_13842559 | 0.12 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr22_+_43011247 | 0.11 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr6_-_161695074 | 0.11 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chrX_-_77225135 | 0.11 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr6_+_106988986 | 0.11 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr16_+_12059091 | 0.11 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr16_+_15489629 | 0.11 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr1_-_160549235 | 0.11 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr12_-_7596735 | 0.11 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr1_-_205091115 | 0.11 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr17_+_25799008 | 0.11 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr17_+_8191815 | 0.10 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr2_-_14541060 | 0.10 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr1_+_8378140 | 0.10 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr11_+_2405833 | 0.10 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr8_-_38008783 | 0.10 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr11_-_128894053 | 0.10 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chrX_-_15288154 | 0.10 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr14_-_54425475 | 0.10 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_76251912 | 0.10 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr17_+_6918093 | 0.10 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr12_-_118796910 | 0.10 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr18_+_29027696 | 0.09 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr21_-_15583165 | 0.09 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr19_-_14992264 | 0.09 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr11_+_55606228 | 0.09 |
ENST00000378396.1
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16 |
| chr1_+_11333245 | 0.09 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr3_+_186692745 | 0.09 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr18_-_52989525 | 0.09 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_-_20053741 | 0.09 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr16_-_66583701 | 0.09 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr12_-_11287243 | 0.08 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr16_-_66583994 | 0.08 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr11_+_115498761 | 0.08 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr2_-_27712583 | 0.08 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr12_-_10282836 | 0.08 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr16_-_18923035 | 0.08 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_37455536 | 0.08 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr1_-_211307404 | 0.08 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr2_+_208414985 | 0.08 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr1_+_197237352 | 0.08 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr7_-_22862406 | 0.08 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr8_+_39972170 | 0.08 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr17_-_41050716 | 0.08 |
ENST00000417193.1
ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671
|
long intergenic non-protein coding RNA 671 |
| chr2_-_165630264 | 0.08 |
ENST00000452626.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr10_-_75415825 | 0.08 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr6_+_26365443 | 0.08 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr7_-_139763521 | 0.08 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_+_100784231 | 0.08 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chrX_-_55057403 | 0.08 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr5_+_140593509 | 0.08 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr19_-_35264089 | 0.08 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr2_-_109605663 | 0.08 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr9_+_93759317 | 0.07 |
ENST00000563268.1
|
RP11-367F23.2
|
RP11-367F23.2 |
| chr12_+_133066137 | 0.07 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr10_-_49860525 | 0.07 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr9_-_5833027 | 0.07 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr16_-_67217844 | 0.07 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr5_+_60933634 | 0.07 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr3_-_145940214 | 0.07 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr18_+_61445205 | 0.07 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr17_+_74463650 | 0.07 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr11_-_26593649 | 0.07 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr18_+_616672 | 0.07 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr4_-_112993808 | 0.07 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr19_-_43969796 | 0.07 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr1_+_233086326 | 0.07 |
ENST00000366628.5
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr19_-_44388116 | 0.07 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr7_+_138943265 | 0.06 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr8_+_24151553 | 0.06 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr10_+_17270214 | 0.06 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr12_+_59194154 | 0.06 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr10_+_52152766 | 0.06 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr17_-_64225508 | 0.06 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_-_71148413 | 0.06 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr19_+_16940198 | 0.06 |
ENST00000248054.5
ENST00000596802.1 ENST00000379803.1 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr11_-_104827425 | 0.06 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_110300607 | 0.06 |
ENST00000260270.2
|
FDX1
|
ferredoxin 1 |
| chr17_+_61086917 | 0.06 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_+_131958436 | 0.06 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr22_-_43010928 | 0.06 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr12_-_42631529 | 0.06 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr14_-_80697396 | 0.06 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr17_-_72772425 | 0.06 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_+_22696314 | 0.06 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr5_-_55412774 | 0.06 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr20_+_3776371 | 0.06 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chrX_-_110655306 | 0.06 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr8_-_128231299 | 0.06 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr7_-_112758665 | 0.05 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr7_+_116654958 | 0.05 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr16_-_66764119 | 0.05 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr6_-_136788001 | 0.05 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr5_+_111755280 | 0.05 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr1_+_56046710 | 0.05 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr12_-_10282742 | 0.05 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_-_152623231 | 0.05 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr12_-_22063787 | 0.05 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_-_186570679 | 0.05 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_70518941 | 0.05 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr1_+_111682058 | 0.05 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr4_-_153601136 | 0.05 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr19_+_9296279 | 0.05 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr4_+_74347400 | 0.05 |
ENST00000226355.3
|
AFM
|
afamin |
| chr2_+_11696464 | 0.05 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr4_+_3344141 | 0.05 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr2_+_172309634 | 0.05 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr14_+_61449197 | 0.04 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr10_+_99627889 | 0.04 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr1_+_162039558 | 0.04 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr13_+_24144796 | 0.04 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_144435985 | 0.04 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr9_-_21482312 | 0.04 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr13_+_32313658 | 0.04 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr6_-_161695042 | 0.04 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.4 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0061150 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.0 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.3 | GO:0034310 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.0 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |