Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
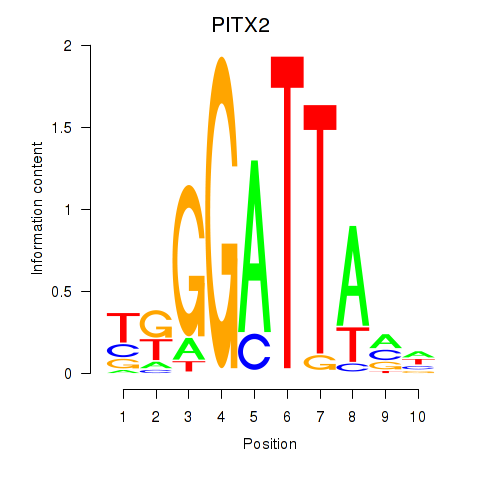
Results for PITX2
Z-value: 0.14
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.11 | paired like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg19_v2_chr4_-_111563279_111563338 | 0.91 | 8.9e-02 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_6949964 | 0.10 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr3_+_119421849 | 0.04 |
ENST00000273390.5
ENST00000463700.1 |
MAATS1
|
MYCBP-associated, testis expressed 1 |
| chr1_+_2407754 | 0.04 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr15_-_89764929 | 0.04 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr9_-_98268883 | 0.04 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr17_-_10633535 | 0.03 |
ENST00000341871.3
|
TMEM220
|
transmembrane protein 220 |
| chr4_+_146560245 | 0.03 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chrM_+_9207 | 0.03 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr21_-_34915084 | 0.03 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr13_+_73632897 | 0.03 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr5_+_64920543 | 0.03 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr3_+_139062838 | 0.03 |
ENST00000310776.4
ENST00000465056.1 ENST00000465373.1 |
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr4_-_76912070 | 0.02 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr12_-_323689 | 0.02 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr4_-_68620053 | 0.02 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr13_-_36429763 | 0.02 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr1_-_31230650 | 0.02 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr12_+_7941989 | 0.02 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr18_-_47017956 | 0.02 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr7_+_73868220 | 0.02 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr7_+_73868439 | 0.02 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr4_+_70916119 | 0.02 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr15_+_41221536 | 0.02 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr6_+_43215727 | 0.02 |
ENST00000304139.5
|
TTBK1
|
tau tubulin kinase 1 |
| chr9_+_34458851 | 0.02 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr3_-_191000172 | 0.02 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr22_-_30234218 | 0.02 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr9_+_34458771 | 0.02 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr2_-_65659762 | 0.01 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr12_-_53189892 | 0.01 |
ENST00000309505.3
ENST00000417996.2 |
KRT3
|
keratin 3 |
| chr4_-_36245561 | 0.01 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_-_111804905 | 0.01 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr21_-_34915147 | 0.01 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr9_+_8858102 | 0.01 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr19_+_2476116 | 0.01 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr16_-_12070468 | 0.01 |
ENST00000538896.1
|
RP11-166B2.1
|
Putative NPIP-like protein LOC729978 |
| chr16_+_28722684 | 0.01 |
ENST00000331666.6
ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chrX_-_21676442 | 0.01 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr2_-_121223697 | 0.01 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr12_-_90049878 | 0.01 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_+_130339710 | 0.01 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_+_120770686 | 0.01 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr11_-_119217365 | 0.01 |
ENST00000360167.4
ENST00000555262.1 ENST00000449574.2 ENST00000445041.2 |
MFRP
C1QTNF5
|
membrane frizzled-related protein C1q and tumor necrosis factor related protein 5 |
| chr5_-_64920115 | 0.01 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr8_-_42360015 | 0.01 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr17_+_4643337 | 0.00 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr6_-_138893661 | 0.00 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr2_+_3622893 | 0.00 |
ENST00000407445.3
ENST00000403564.1 |
RPS7
|
ribosomal protein S7 |
| chr1_-_98515395 | 0.00 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr6_+_35310391 | 0.00 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr8_-_10512569 | 0.00 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr2_+_120770645 | 0.00 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr17_+_4643300 | 0.00 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr6_-_41673552 | 0.00 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr17_-_4643114 | 0.00 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
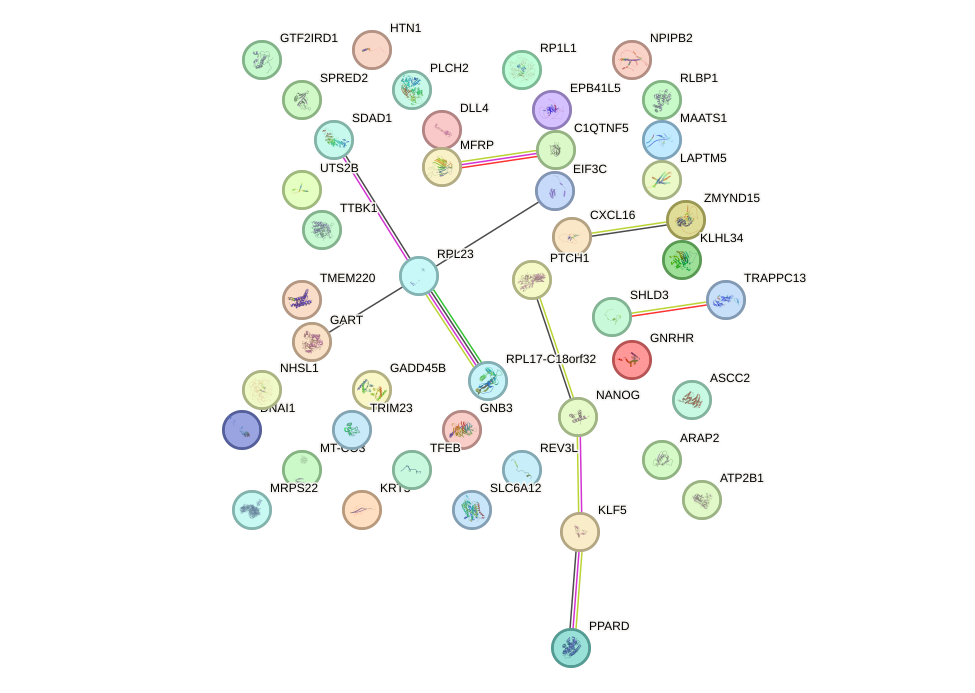
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.0 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |