Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
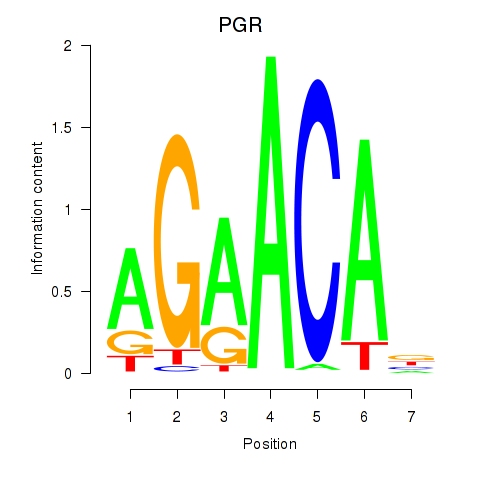
Results for PGR
Z-value: 0.58
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_100999775_100999801 | -0.44 | 5.6e-01 | Click! |
Activity profile of PGR motif
Sorted Z-values of PGR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_148442844 | 0.34 |
ENST00000515519.1
|
CTC-529P8.1
|
CTC-529P8.1 |
| chrX_+_109602039 | 0.30 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr2_-_220034745 | 0.25 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr16_+_56970567 | 0.24 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr1_-_145715565 | 0.22 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr11_+_35222629 | 0.22 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr4_-_88450612 | 0.21 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr13_+_102104980 | 0.20 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr9_+_82188077 | 0.20 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_-_25451065 | 0.19 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr2_-_27531313 | 0.18 |
ENST00000296099.2
|
UCN
|
urocortin |
| chr14_-_61124977 | 0.17 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr3_+_52454971 | 0.17 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chrX_-_100641155 | 0.16 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr3_-_124839648 | 0.16 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr6_+_27107053 | 0.16 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr7_-_75241096 | 0.16 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr7_+_22766766 | 0.16 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr7_-_37026108 | 0.15 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr10_-_28571015 | 0.15 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr15_+_31658349 | 0.15 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr19_+_41119794 | 0.15 |
ENST00000593463.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr2_+_232135245 | 0.15 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr2_+_38177575 | 0.15 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr17_-_27188984 | 0.15 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr19_-_54726850 | 0.14 |
ENST00000245620.9
ENST00000346401.6 ENST00000424807.1 ENST00000445347.1 |
LILRB3
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
| chr11_+_65627974 | 0.14 |
ENST00000525768.1
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr11_+_3875757 | 0.14 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr2_+_173686303 | 0.14 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr17_-_26697304 | 0.14 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr20_-_25207370 | 0.13 |
ENST00000593352.1
|
AL035252.1
|
HCG2018895; Uncharacterized protein |
| chr19_-_41903161 | 0.13 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr11_+_65999265 | 0.13 |
ENST00000528935.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr2_-_158182105 | 0.13 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr11_+_55606228 | 0.12 |
ENST00000378396.1
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16 |
| chr7_+_99202003 | 0.12 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr4_-_89442940 | 0.12 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr2_-_220034712 | 0.12 |
ENST00000409370.2
ENST00000430764.1 ENST00000409878.3 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr3_+_49057876 | 0.12 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr2_+_9778872 | 0.12 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr8_+_145065705 | 0.12 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr14_-_75330537 | 0.12 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr16_-_28506840 | 0.11 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr6_-_32908765 | 0.11 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr19_+_35773242 | 0.11 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr12_+_56661461 | 0.11 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr5_+_52776449 | 0.11 |
ENST00000396947.3
|
FST
|
follistatin |
| chr1_-_179834311 | 0.10 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr2_-_158182322 | 0.10 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr17_-_39681578 | 0.10 |
ENST00000593096.1
|
KRT19
|
keratin 19 |
| chr1_+_155583012 | 0.10 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr19_+_13135439 | 0.10 |
ENST00000586873.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr9_-_127710292 | 0.10 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr22_-_31688381 | 0.10 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_-_73693875 | 0.09 |
ENST00000536983.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr17_-_4463856 | 0.09 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr19_+_30863271 | 0.09 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr11_-_65625014 | 0.09 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr4_-_23735183 | 0.09 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr17_+_46970178 | 0.09 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_22022437 | 0.09 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr10_+_90562487 | 0.09 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr2_+_42104692 | 0.09 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr9_+_71394945 | 0.08 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr22_-_31688431 | 0.08 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr10_+_63808970 | 0.08 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr17_-_17480779 | 0.08 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chrX_+_54834159 | 0.08 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr2_-_188312971 | 0.08 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr6_-_43276535 | 0.08 |
ENST00000372569.3
ENST00000274990.4 |
CRIP3
|
cysteine-rich protein 3 |
| chr10_+_90562705 | 0.08 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr17_-_26903900 | 0.08 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr13_+_24144509 | 0.08 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr22_-_30162924 | 0.08 |
ENST00000344318.3
ENST00000397781.3 |
ZMAT5
|
zinc finger, matrin-type 5 |
| chr5_+_102200948 | 0.08 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_227771404 | 0.08 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr1_+_192778161 | 0.07 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr7_-_5821225 | 0.07 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr19_+_41119323 | 0.07 |
ENST00000599724.1
ENST00000597071.1 ENST00000243562.9 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr1_-_205290865 | 0.07 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr4_-_105416039 | 0.07 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr11_-_66725837 | 0.07 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr7_-_14026123 | 0.07 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr16_-_20338748 | 0.07 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr2_+_121011027 | 0.07 |
ENST00000449649.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr13_+_78315295 | 0.07 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_41174988 | 0.07 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr4_+_177241094 | 0.07 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr12_-_69326940 | 0.07 |
ENST00000549781.1
ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM
|
carboxypeptidase M |
| chr11_-_10920838 | 0.07 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr8_+_35649365 | 0.07 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr14_+_24674926 | 0.07 |
ENST00000339917.5
ENST00000556621.1 ENST00000287913.6 ENST00000428351.2 ENST00000555092.1 |
TSSK4
|
testis-specific serine kinase 4 |
| chr3_+_25469724 | 0.07 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr3_-_49722523 | 0.06 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr8_+_145065521 | 0.06 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr15_-_74726283 | 0.06 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr16_-_3306587 | 0.06 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr19_+_51153045 | 0.06 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr2_+_130737223 | 0.06 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr9_+_131683174 | 0.06 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr3_+_137906353 | 0.06 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr12_-_122238464 | 0.06 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr3_+_137906154 | 0.06 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr2_+_191208601 | 0.06 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr5_+_139505520 | 0.06 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr6_+_30951487 | 0.06 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr3_+_137906109 | 0.06 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr17_+_41150793 | 0.06 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr17_+_25958174 | 0.06 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chrX_+_47093171 | 0.06 |
ENST00000377078.2
|
USP11
|
ubiquitin specific peptidase 11 |
| chr19_+_507299 | 0.06 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr16_+_57679859 | 0.06 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_26694979 | 0.06 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr7_-_14026063 | 0.06 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr6_-_32098013 | 0.06 |
ENST00000375156.3
|
FKBPL
|
FK506 binding protein like |
| chr6_-_3912207 | 0.05 |
ENST00000566733.1
|
RP1-140K8.5
|
RP1-140K8.5 |
| chr12_+_54384370 | 0.05 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr17_+_60501228 | 0.05 |
ENST00000311506.5
|
METTL2A
|
methyltransferase like 2A |
| chr8_+_11534462 | 0.05 |
ENST00000528712.1
ENST00000532977.1 |
GATA4
|
GATA binding protein 4 |
| chr16_+_57680811 | 0.05 |
ENST00000569101.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr5_-_134735568 | 0.05 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr8_+_22446763 | 0.05 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr12_+_122242597 | 0.05 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr3_+_41236325 | 0.05 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr2_+_163175394 | 0.05 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr12_+_21654714 | 0.05 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr2_-_179315786 | 0.05 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr11_-_75379612 | 0.05 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr12_+_50146747 | 0.05 |
ENST00000547798.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr13_+_78315528 | 0.05 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr17_+_73997796 | 0.05 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr10_+_124030819 | 0.05 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr1_+_155146318 | 0.05 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr12_-_88974236 | 0.05 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr6_-_31704282 | 0.05 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr4_+_79567314 | 0.05 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chrX_-_45060135 | 0.05 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr17_+_41150479 | 0.05 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr1_+_155023757 | 0.05 |
ENST00000356955.2
ENST00000449910.2 ENST00000359280.4 ENST00000360674.4 ENST00000368412.3 ENST00000355956.2 ENST00000368410.2 ENST00000271836.6 ENST00000368413.1 ENST00000531455.1 ENST00000447332.3 |
ADAM15
|
ADAM metallopeptidase domain 15 |
| chr5_+_66124590 | 0.05 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_129845313 | 0.05 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr17_-_42466864 | 0.05 |
ENST00000353281.4
ENST00000262407.5 |
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr16_+_57680840 | 0.05 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_16010779 | 0.05 |
ENST00000375799.3
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr3_+_25469802 | 0.04 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr2_-_134326009 | 0.04 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr15_+_36887069 | 0.04 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_+_41447609 | 0.04 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chrX_+_16964985 | 0.04 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr5_+_138629337 | 0.04 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr9_-_140196703 | 0.04 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chrX_-_69509738 | 0.04 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr20_+_34129770 | 0.04 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr5_+_133861790 | 0.04 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr5_+_140261703 | 0.04 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr22_+_42017987 | 0.04 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr19_+_49377575 | 0.04 |
ENST00000600406.1
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr6_-_32908792 | 0.04 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr2_-_165698322 | 0.04 |
ENST00000444537.1
ENST00000414843.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chrX_-_153599578 | 0.04 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr20_+_35090150 | 0.04 |
ENST00000340491.4
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr10_+_69865866 | 0.04 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr2_+_105050794 | 0.04 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr12_+_131438443 | 0.04 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chrX_+_54834004 | 0.04 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr1_+_145516560 | 0.04 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr2_+_27440229 | 0.04 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr17_+_39411636 | 0.04 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chrX_-_132095419 | 0.04 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr16_+_57679945 | 0.04 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr2_-_85641162 | 0.04 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr2_+_102508955 | 0.04 |
ENST00000414004.2
|
FLJ20373
|
FLJ20373 |
| chr5_-_64064508 | 0.04 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr1_+_160321120 | 0.03 |
ENST00000424754.1
|
NCSTN
|
nicastrin |
| chr5_-_180242534 | 0.03 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr2_+_219536749 | 0.03 |
ENST00000295709.3
ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36
|
serine/threonine kinase 36 |
| chr3_+_42695176 | 0.03 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr10_-_72142345 | 0.03 |
ENST00000373224.1
ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20
|
leucine rich repeat containing 20 |
| chr20_-_25320367 | 0.03 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr11_+_57105991 | 0.03 |
ENST00000263314.2
|
P2RX3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr6_+_143447322 | 0.03 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr17_+_65027509 | 0.03 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr2_+_99797542 | 0.03 |
ENST00000338148.3
ENST00000512183.2 |
MRPL30
C2orf15
|
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr11_-_65625330 | 0.03 |
ENST00000531407.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr7_+_73442457 | 0.03 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr17_+_79031415 | 0.03 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr17_-_27467418 | 0.03 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr2_+_191208196 | 0.03 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_+_212475148 | 0.03 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr2_+_219537015 | 0.03 |
ENST00000440309.1
ENST00000424080.1 |
STK36
|
serine/threonine kinase 36 |
| chr18_-_53019208 | 0.03 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr19_+_16771936 | 0.03 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr2_-_165697717 | 0.03 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr15_+_65843130 | 0.03 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr11_-_6502534 | 0.03 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr9_-_37034028 | 0.03 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr7_-_100491854 | 0.03 |
ENST00000426415.1
ENST00000430554.1 ENST00000412389.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr9_-_131940526 | 0.03 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr1_-_201390846 | 0.03 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr19_-_8642289 | 0.03 |
ENST00000596675.1
ENST00000338257.8 |
MYO1F
|
myosin IF |
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.2 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) |
| 0.1 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.0 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:2000769 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:1902310 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |