Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
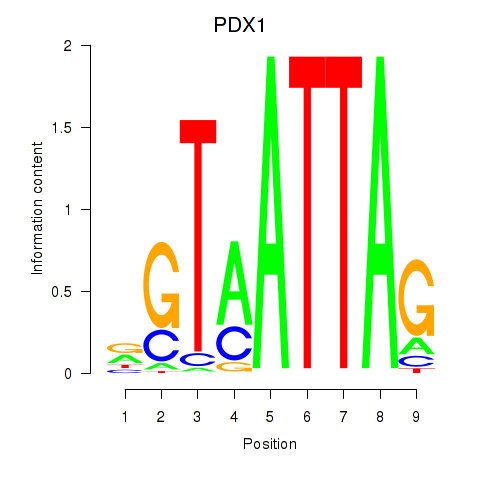
Results for PDX1
Z-value: 0.73
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | -0.16 | 8.4e-01 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_46690839 | 0.44 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr7_-_99716914 | 0.31 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_66918558 | 0.29 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr1_-_149859466 | 0.26 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr6_-_116833500 | 0.24 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr7_+_116660246 | 0.23 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr3_-_167191814 | 0.20 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr15_+_63188009 | 0.20 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr5_-_56778635 | 0.19 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr18_+_57567180 | 0.19 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr20_-_31124186 | 0.19 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr1_+_198608146 | 0.18 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr17_-_57229155 | 0.18 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_+_115498761 | 0.18 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr11_+_35222629 | 0.18 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_62439037 | 0.15 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr21_+_17443521 | 0.14 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_144354644 | 0.14 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chrX_+_133930798 | 0.14 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr21_+_29911640 | 0.12 |
ENST00000412526.1
ENST00000455939.1 |
LINC00161
|
long intergenic non-protein coding RNA 161 |
| chr2_+_45168875 | 0.12 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr18_-_33709268 | 0.12 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr19_+_13049413 | 0.12 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr3_-_98235962 | 0.11 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr8_-_37594944 | 0.11 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr20_-_271304 | 0.11 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr1_+_28261533 | 0.11 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr4_+_146402346 | 0.10 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr14_-_36988882 | 0.10 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr7_+_6655225 | 0.10 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr17_-_2117600 | 0.10 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr5_-_134735568 | 0.09 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr2_-_190927447 | 0.09 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chrX_+_72783026 | 0.09 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr4_-_103749105 | 0.09 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr21_+_17443434 | 0.09 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_-_123639304 | 0.09 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr9_+_131062367 | 0.08 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr2_-_99871570 | 0.08 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr15_-_55562582 | 0.08 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_-_57233966 | 0.08 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr13_+_78315528 | 0.08 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr11_-_31531121 | 0.08 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chrX_-_133931164 | 0.08 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr9_+_131037623 | 0.08 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr2_+_191208791 | 0.08 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr9_-_16728161 | 0.08 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr15_+_43886057 | 0.08 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr15_-_55562479 | 0.07 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_+_74846230 | 0.07 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr17_-_8301132 | 0.07 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr12_+_64798826 | 0.07 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr17_+_18086392 | 0.07 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr13_+_102104980 | 0.07 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr15_+_43986069 | 0.07 |
ENST00000415044.1
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr7_-_14028488 | 0.07 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr2_-_145277569 | 0.06 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_-_103749313 | 0.06 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_56325231 | 0.06 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr15_-_31393910 | 0.06 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr18_+_32558208 | 0.06 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr11_+_112041253 | 0.06 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr14_-_61116168 | 0.06 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr21_-_35899113 | 0.06 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr3_+_111717511 | 0.06 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr6_-_38670897 | 0.06 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr12_-_86650077 | 0.06 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_+_80351977 | 0.06 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr2_-_136678123 | 0.06 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr9_-_136933615 | 0.06 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr4_+_141445311 | 0.06 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr4_+_86525299 | 0.06 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_-_96478466 | 0.06 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_+_66300446 | 0.06 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr18_-_47792851 | 0.06 |
ENST00000398545.4
|
CCDC11
|
coiled-coil domain containing 11 |
| chrX_-_64196307 | 0.05 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr12_-_54813229 | 0.05 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr21_+_17442799 | 0.05 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_+_90012986 | 0.05 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chrX_+_591524 | 0.05 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr3_-_108248169 | 0.05 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr4_+_95128748 | 0.05 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_+_65554493 | 0.05 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr9_-_136933134 | 0.05 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr2_+_48541776 | 0.05 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr1_-_1710229 | 0.05 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr13_-_31191642 | 0.05 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr1_-_232651312 | 0.05 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr15_+_69854027 | 0.05 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr3_-_112127981 | 0.05 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr17_-_38928414 | 0.05 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr14_-_54423529 | 0.05 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr9_-_123639445 | 0.05 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chrX_-_19817869 | 0.04 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_+_45241109 | 0.04 |
ENST00000396651.3
|
RPS8
|
ribosomal protein S8 |
| chr13_-_99667960 | 0.04 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr15_+_80351910 | 0.04 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr1_+_212965170 | 0.04 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr12_-_56105880 | 0.04 |
ENST00000557257.1
|
ITGA7
|
integrin, alpha 7 |
| chr12_-_53729525 | 0.04 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr1_+_36621174 | 0.04 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr2_+_191208601 | 0.04 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr11_-_85430204 | 0.04 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_+_28586006 | 0.04 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chrX_-_106243451 | 0.04 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr3_+_111717600 | 0.04 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr18_+_3447572 | 0.04 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_-_130423200 | 0.04 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr17_-_39623681 | 0.04 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr15_+_59279851 | 0.04 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr1_+_84609944 | 0.04 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_+_43985725 | 0.04 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_+_151735431 | 0.04 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr4_+_71587669 | 0.04 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr12_-_14849470 | 0.04 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr6_+_39760129 | 0.04 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_+_56325812 | 0.04 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr4_+_77356248 | 0.04 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr15_+_34261089 | 0.04 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr15_-_37393406 | 0.04 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr3_+_111718036 | 0.04 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr14_+_39944025 | 0.04 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr3_-_39321512 | 0.03 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr12_-_56321397 | 0.03 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr12_-_94673956 | 0.03 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr6_+_150920999 | 0.03 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_94147385 | 0.03 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_135379132 | 0.03 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chrX_-_106243294 | 0.03 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr3_-_121740969 | 0.03 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr12_+_49621658 | 0.03 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr2_+_196313239 | 0.03 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr1_+_66796401 | 0.03 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr4_-_41884620 | 0.03 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr3_+_69985734 | 0.03 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chrX_-_110655306 | 0.03 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr19_+_36632204 | 0.03 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr20_-_50419055 | 0.03 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr4_+_53525573 | 0.03 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr7_-_27169801 | 0.03 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr18_-_46784778 | 0.03 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr7_-_99717463 | 0.03 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_-_39979576 | 0.03 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr10_+_118083919 | 0.03 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr18_+_32558380 | 0.03 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_92247032 | 0.03 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr2_+_109204743 | 0.03 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_38956205 | 0.03 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr18_-_44181442 | 0.03 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr12_-_57328187 | 0.03 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr5_+_66300464 | 0.03 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_-_38071122 | 0.03 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr6_+_13272904 | 0.03 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr6_-_111136513 | 0.03 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr19_+_11071546 | 0.03 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr15_+_62853562 | 0.02 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr12_-_56106060 | 0.02 |
ENST00000452168.2
|
ITGA7
|
integrin, alpha 7 |
| chr2_-_61697862 | 0.02 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr10_+_119302508 | 0.02 |
ENST00000442245.4
|
EMX2
|
empty spiracles homeobox 2 |
| chr17_+_60536002 | 0.02 |
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2 |
| chr6_+_167704838 | 0.02 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr8_-_1922789 | 0.02 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr7_+_1272522 | 0.02 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr6_-_167040693 | 0.02 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chrX_-_64196376 | 0.02 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr5_+_135394840 | 0.02 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr20_-_49308048 | 0.02 |
ENST00000327979.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr2_+_191208656 | 0.02 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr4_-_155533787 | 0.02 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chrX_+_7137475 | 0.02 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr13_-_46716969 | 0.02 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_-_1709845 | 0.02 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr7_-_33102338 | 0.02 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr6_-_100912785 | 0.02 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr12_-_53730126 | 0.02 |
ENST00000537210.2
|
SP7
|
Sp7 transcription factor |
| chr15_-_65903407 | 0.02 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr1_+_159272111 | 0.02 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr14_+_59100774 | 0.02 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr11_+_118403747 | 0.02 |
ENST00000526853.1
|
TMEM25
|
transmembrane protein 25 |
| chr5_-_140998481 | 0.02 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr5_+_156712372 | 0.02 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_+_114710211 | 0.02 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr21_+_17792672 | 0.02 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_+_125034586 | 0.02 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr8_+_92261516 | 0.02 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr1_-_169396646 | 0.02 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr9_-_124990680 | 0.02 |
ENST00000541397.2
ENST00000560485.1 |
LHX6
|
LIM homeobox 6 |
| chrX_-_64196351 | 0.02 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr4_-_103748880 | 0.02 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_151431909 | 0.02 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr12_-_53730147 | 0.02 |
ENST00000536324.2
|
SP7
|
Sp7 transcription factor |
| chr17_-_4458616 | 0.02 |
ENST00000381556.2
|
MYBBP1A
|
MYB binding protein (P160) 1a |
| chr18_+_55888767 | 0.02 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_130318869 | 0.02 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr1_+_68150744 | 0.02 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr1_-_10532531 | 0.01 |
ENST00000377036.2
ENST00000377038.3 |
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr7_-_81399438 | 0.01 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr4_-_152147579 | 0.01 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr14_-_77889860 | 0.01 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |