Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
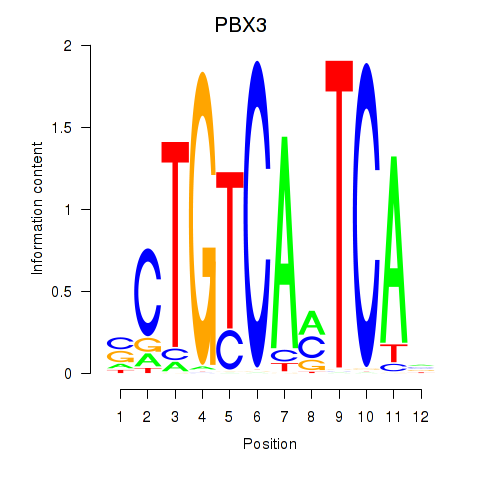
Results for PBX3
Z-value: 0.41
Transcription factors associated with PBX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX3
|
ENSG00000167081.12 | PBX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX3 | hg19_v2_chr9_+_128509624_128509658 | -0.85 | 1.5e-01 | Click! |
Activity profile of PBX3 motif
Sorted Z-values of PBX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_79369249 | 0.54 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr1_-_12679171 | 0.24 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr19_-_23578220 | 0.19 |
ENST00000595533.1
ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91
|
zinc finger protein 91 |
| chr6_-_30685214 | 0.16 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr7_-_44105158 | 0.15 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chrX_-_153279697 | 0.15 |
ENST00000444254.1
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr18_+_44526786 | 0.14 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr17_+_61699766 | 0.14 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr14_-_75530693 | 0.14 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr11_+_125439176 | 0.13 |
ENST00000529812.1
|
EI24
|
etoposide induced 2.4 |
| chr12_-_123717711 | 0.13 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr19_-_12662314 | 0.12 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr2_+_220094657 | 0.12 |
ENST00000436226.1
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr14_+_23790655 | 0.12 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr8_-_101964738 | 0.12 |
ENST00000523938.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr20_-_35807741 | 0.12 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr4_-_140477910 | 0.12 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr17_+_46125685 | 0.12 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr17_+_38376042 | 0.12 |
ENST00000583130.1
ENST00000584296.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr5_+_2752216 | 0.12 |
ENST00000457752.2
|
C5orf38
|
chromosome 5 open reading frame 38 |
| chr5_-_176057518 | 0.11 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr1_+_85527987 | 0.11 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr22_-_39151463 | 0.10 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr12_-_52604607 | 0.10 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr8_-_57123815 | 0.10 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr22_-_39151995 | 0.10 |
ENST00000405018.1
ENST00000438058.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr4_-_13546632 | 0.09 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr19_-_49140692 | 0.09 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_-_41870026 | 0.09 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr5_-_81046904 | 0.09 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr5_+_150406527 | 0.09 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr17_+_58755184 | 0.08 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr1_-_935491 | 0.08 |
ENST00000304952.6
|
HES4
|
hes family bHLH transcription factor 4 |
| chr9_-_98079154 | 0.08 |
ENST00000433829.1
|
FANCC
|
Fanconi anemia, complementation group C |
| chr5_+_121647877 | 0.08 |
ENST00000514497.2
ENST00000261367.7 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr4_-_140477353 | 0.08 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr11_-_4629367 | 0.08 |
ENST00000533021.1
|
TRIM68
|
tripartite motif containing 68 |
| chr2_+_42275153 | 0.08 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr19_-_47551836 | 0.08 |
ENST00000253047.6
|
TMEM160
|
transmembrane protein 160 |
| chr10_+_67330096 | 0.08 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr10_-_76868931 | 0.08 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr19_+_16435625 | 0.08 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr19_-_49140609 | 0.08 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr22_-_45559540 | 0.08 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr8_-_101964832 | 0.07 |
ENST00000523131.1
ENST00000418997.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr10_+_114710516 | 0.07 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_+_59783941 | 0.07 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr5_-_138730817 | 0.07 |
ENST00000434752.2
|
PROB1
|
proline-rich basic protein 1 |
| chr22_-_39151947 | 0.07 |
ENST00000216064.4
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr17_-_1619568 | 0.07 |
ENST00000571595.1
|
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr12_+_51632666 | 0.07 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr19_-_21512115 | 0.07 |
ENST00000601295.1
|
ZNF708
|
zinc finger protein 708 |
| chr17_-_56591978 | 0.07 |
ENST00000583656.1
|
MTMR4
|
myotubularin related protein 4 |
| chr14_-_65769392 | 0.07 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr12_+_28343353 | 0.07 |
ENST00000539107.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr11_+_842928 | 0.07 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr11_-_842509 | 0.07 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr1_-_247171347 | 0.07 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr6_-_31620095 | 0.07 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr10_+_114710425 | 0.07 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_110881945 | 0.07 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr19_+_55591743 | 0.07 |
ENST00000588359.1
ENST00000245618.5 |
EPS8L1
|
EPS8-like 1 |
| chr17_+_19314505 | 0.07 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr3_+_48481658 | 0.07 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr14_+_39644425 | 0.06 |
ENST00000556530.1
|
PNN
|
pinin, desmosome associated protein |
| chr12_+_31227192 | 0.06 |
ENST00000535317.1
|
DDX11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr2_-_220436248 | 0.06 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr2_-_61389168 | 0.06 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr1_+_32674675 | 0.06 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr7_-_38370536 | 0.06 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chr17_-_65992544 | 0.06 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr2_-_98612379 | 0.06 |
ENST00000425805.2
|
TMEM131
|
transmembrane protein 131 |
| chr16_-_29275800 | 0.06 |
ENST00000567984.1
|
RP11-426C22.6
|
RP11-426C22.6 |
| chr9_-_35689900 | 0.06 |
ENST00000378300.5
ENST00000329305.2 ENST00000360958.2 |
TPM2
|
tropomyosin 2 (beta) |
| chr7_-_100183742 | 0.06 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr2_+_63277927 | 0.06 |
ENST00000282549.2
|
OTX1
|
orthodenticle homeobox 1 |
| chrX_-_107681633 | 0.06 |
ENST00000394872.2
ENST00000334504.7 |
COL4A6
|
collagen, type IV, alpha 6 |
| chr19_-_2740036 | 0.06 |
ENST00000586572.1
ENST00000269740.4 |
AC006538.4
SLC39A3
|
Uncharacterized protein solute carrier family 39 (zinc transporter), member 3 |
| chr21_-_30257669 | 0.06 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr7_+_150929550 | 0.06 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr2_+_66662249 | 0.06 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr10_-_22292675 | 0.06 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr4_+_7940728 | 0.06 |
ENST00000429019.3
|
AC097381.1
|
AC097381.1 |
| chr3_+_111393501 | 0.06 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_-_935519 | 0.06 |
ENST00000428771.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chr19_-_20844368 | 0.06 |
ENST00000595094.1
ENST00000601440.1 ENST00000291750.6 |
CTC-513N18.7
ZNF626
|
CTC-513N18.7 zinc finger protein 626 |
| chr2_+_65663812 | 0.06 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr19_-_11266437 | 0.06 |
ENST00000586708.1
ENST00000591396.1 ENST00000592967.1 ENST00000585486.1 ENST00000585567.1 |
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr11_+_842808 | 0.06 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr1_+_230202936 | 0.06 |
ENST00000366672.4
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr19_+_11485333 | 0.06 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr3_+_30648066 | 0.06 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr12_-_109219937 | 0.06 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chrX_+_70503526 | 0.06 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chrX_+_150148976 | 0.06 |
ENST00000419110.1
|
HMGB3
|
high mobility group box 3 |
| chr17_+_38375528 | 0.05 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr20_+_43343476 | 0.05 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr7_+_64254766 | 0.05 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr3_+_107241783 | 0.05 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr10_-_21806759 | 0.05 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr5_-_81046841 | 0.05 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr2_+_113033164 | 0.05 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr11_+_117015024 | 0.05 |
ENST00000530272.1
|
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr4_+_26859300 | 0.05 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chr2_+_232826394 | 0.05 |
ENST00000409401.3
ENST00000441279.1 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr16_+_67927147 | 0.05 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr2_+_74685413 | 0.05 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr3_+_122920847 | 0.05 |
ENST00000466519.1
ENST00000480631.1 ENST00000491366.1 ENST00000487572.1 |
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
| chr3_-_141747950 | 0.05 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_-_175204765 | 0.05 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr3_+_111393659 | 0.05 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_-_156571254 | 0.05 |
ENST00000438976.2
ENST00000334588.7 ENST00000368232.4 ENST00000415314.2 |
GPATCH4
|
G patch domain containing 4 |
| chr5_+_59783540 | 0.05 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chrX_-_107681555 | 0.05 |
ENST00000545689.1
ENST00000538570.1 |
COL4A6
|
collagen, type IV, alpha 6 |
| chr19_+_21106081 | 0.05 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr3_-_186080012 | 0.05 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr13_-_20437648 | 0.05 |
ENST00000382907.4
ENST00000382905.4 |
ZMYM5
|
zinc finger, MYM-type 5 |
| chr12_-_91573132 | 0.05 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr19_+_21324863 | 0.05 |
ENST00000598331.1
|
ZNF431
|
zinc finger protein 431 |
| chrX_-_47930980 | 0.05 |
ENST00000442455.3
ENST00000428686.1 ENST00000276054.4 |
ZNF630
|
zinc finger protein 630 |
| chr19_+_51728316 | 0.05 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr5_-_137514333 | 0.05 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr5_+_2752334 | 0.05 |
ENST00000505778.1
ENST00000515640.1 ENST00000397835.4 |
C5orf38
|
chromosome 5 open reading frame 38 |
| chr11_+_58938903 | 0.05 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr19_-_22193731 | 0.05 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr1_-_935361 | 0.05 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chr12_+_28343365 | 0.05 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr3_+_188817891 | 0.05 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr3_+_30647994 | 0.05 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr12_+_38710555 | 0.05 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr8_-_101965104 | 0.05 |
ENST00000437293.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr12_+_7055631 | 0.05 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr19_-_59084647 | 0.05 |
ENST00000594234.1
ENST00000596039.1 |
MZF1
|
myeloid zinc finger 1 |
| chr2_-_220435963 | 0.05 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr5_+_43602750 | 0.04 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chrX_+_75392771 | 0.04 |
ENST00000373358.3
ENST00000373357.3 |
PBDC1
|
polysaccharide biosynthesis domain containing 1 |
| chr1_+_12538594 | 0.04 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr12_+_51633061 | 0.04 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr3_-_113415441 | 0.04 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr1_+_161195835 | 0.04 |
ENST00000545897.1
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr3_+_101498026 | 0.04 |
ENST00000495842.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr16_+_1756162 | 0.04 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr13_+_20207782 | 0.04 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chrX_-_80377162 | 0.04 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr17_+_48423453 | 0.04 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr3_+_156807663 | 0.04 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr1_+_164528437 | 0.04 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr16_+_1728305 | 0.04 |
ENST00000569765.1
|
HN1L
|
hematological and neurological expressed 1-like |
| chr7_+_75024903 | 0.04 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr13_-_79177673 | 0.04 |
ENST00000377208.5
|
POU4F1
|
POU class 4 homeobox 1 |
| chr2_-_61389240 | 0.04 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr10_+_74451883 | 0.04 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr3_+_52570610 | 0.04 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr10_+_6779326 | 0.04 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr19_+_4247070 | 0.04 |
ENST00000262962.7
|
CCDC94
|
coiled-coil domain containing 94 |
| chr19_+_49375649 | 0.04 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr8_-_93978216 | 0.04 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr5_-_176057365 | 0.04 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr4_-_2420357 | 0.04 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr19_+_4969116 | 0.04 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr19_+_13906250 | 0.04 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr9_+_119449576 | 0.04 |
ENST00000450136.1
ENST00000373983.2 ENST00000411410.1 |
TRIM32
|
tripartite motif containing 32 |
| chr3_+_52279737 | 0.04 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr7_+_64838712 | 0.04 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr15_-_81282133 | 0.04 |
ENST00000261758.4
|
MESDC2
|
mesoderm development candidate 2 |
| chr19_+_49999631 | 0.04 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr1_+_17531614 | 0.04 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chrX_+_69674943 | 0.04 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr4_+_331578 | 0.04 |
ENST00000512994.1
|
ZNF141
|
zinc finger protein 141 |
| chr17_+_38375574 | 0.04 |
ENST00000323571.4
ENST00000585043.1 ENST00000394103.3 ENST00000536600.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr10_+_114710211 | 0.04 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_-_46916805 | 0.04 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr6_-_111888474 | 0.04 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr12_-_118406777 | 0.04 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr5_-_137514617 | 0.04 |
ENST00000254900.5
|
BRD8
|
bromodomain containing 8 |
| chr3_+_14474178 | 0.04 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr7_-_148725544 | 0.04 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chrX_-_80377118 | 0.04 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chrX_+_15767971 | 0.04 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr4_-_175443788 | 0.04 |
ENST00000541923.1
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr19_-_21512202 | 0.04 |
ENST00000356929.3
|
ZNF708
|
zinc finger protein 708 |
| chr1_-_106161540 | 0.04 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr16_+_4382225 | 0.04 |
ENST00000433375.1
|
GLIS2
|
GLIS family zinc finger 2 |
| chr6_-_94129244 | 0.04 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr1_+_174968564 | 0.04 |
ENST00000426793.1
|
CACYBP
|
calcyclin binding protein |
| chr12_-_42983478 | 0.04 |
ENST00000345127.3
ENST00000547113.1 |
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr17_+_46125707 | 0.04 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr7_-_14029283 | 0.04 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr1_+_22778337 | 0.03 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr6_+_36853607 | 0.03 |
ENST00000480824.2
ENST00000355190.3 ENST00000373685.1 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr17_-_44657017 | 0.03 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr14_-_73925225 | 0.03 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr19_-_20748614 | 0.03 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr1_-_156470515 | 0.03 |
ENST00000340875.5
ENST00000368240.2 ENST00000353795.3 |
MEF2D
|
myocyte enhancer factor 2D |
| chr15_+_41221536 | 0.03 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr19_-_2739992 | 0.03 |
ENST00000545664.1
ENST00000589363.1 ENST00000455372.2 |
SLC39A3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr12_+_51632638 | 0.03 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr1_+_6845384 | 0.03 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr1_-_22109682 | 0.03 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr2_-_176033066 | 0.03 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
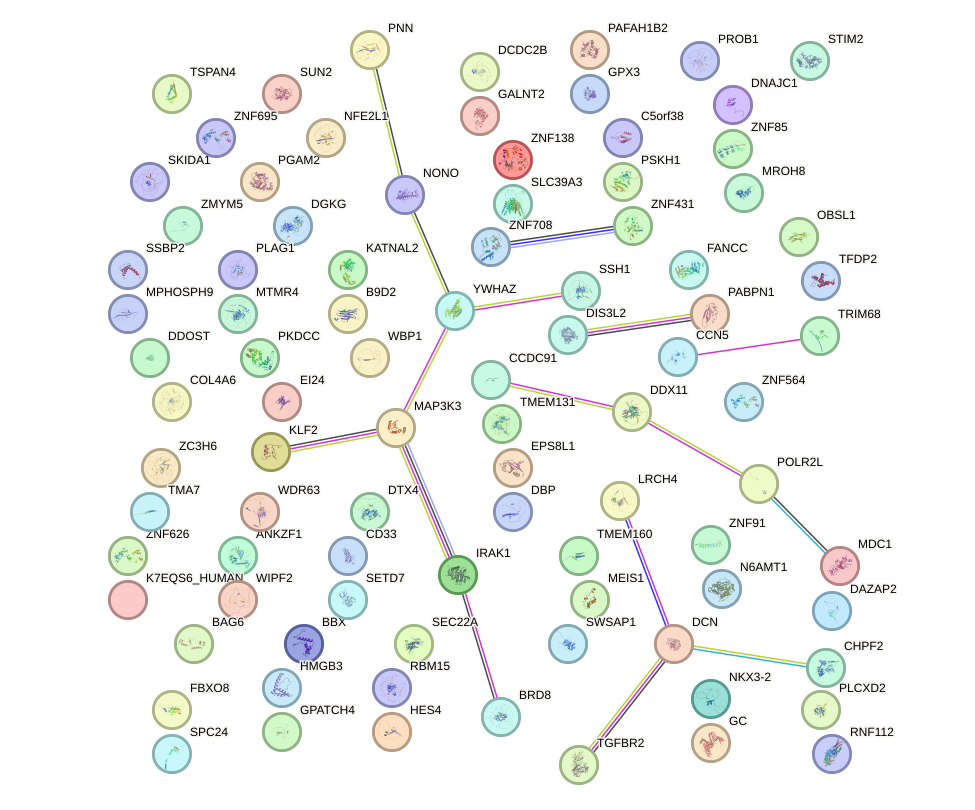
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.0 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |