Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
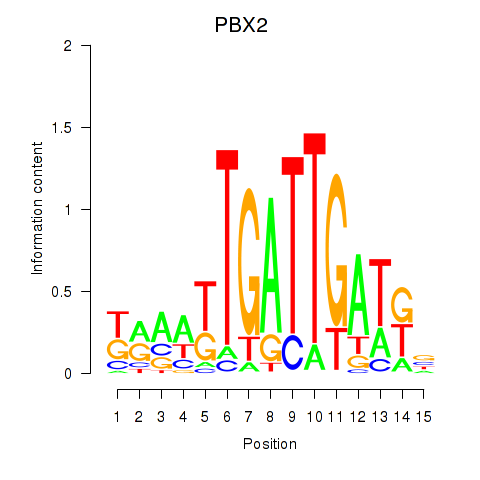
Results for PBX2
Z-value: 0.11
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.7 | PBX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX2 | hg19_v2_chr6_-_32157947_32157992 | 0.78 | 2.2e-01 | Click! |
Activity profile of PBX2 motif
Sorted Z-values of PBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_54317532 | 0.04 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr9_-_20382446 | 0.04 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr3_+_159943362 | 0.03 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr12_-_28125638 | 0.03 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr2_-_99917639 | 0.02 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr17_+_7758374 | 0.02 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr11_-_62432641 | 0.02 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr4_-_100065440 | 0.02 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr16_+_1128781 | 0.02 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr12_-_28124903 | 0.02 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr14_-_55658323 | 0.02 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr10_+_47894572 | 0.02 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr13_-_107220455 | 0.02 |
ENST00000400198.3
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr14_-_55658252 | 0.01 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr2_+_33661382 | 0.01 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_-_75008244 | 0.01 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr14_+_58765305 | 0.01 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_134528635 | 0.01 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr5_-_39462390 | 0.01 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_-_120189900 | 0.01 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr2_+_113033164 | 0.01 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr17_-_29151686 | 0.01 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr2_-_190044480 | 0.01 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr1_-_26231589 | 0.01 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chrX_+_119737806 | 0.01 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr14_+_38033252 | 0.01 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr11_-_104769141 | 0.01 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr12_+_53835425 | 0.01 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr7_-_33140498 | 0.01 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr14_+_58765103 | 0.01 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chrM_+_12331 | 0.01 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr3_-_52864680 | 0.00 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr15_+_67418047 | 0.00 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr3_+_129158926 | 0.00 |
ENST00000347300.2
ENST00000296266.3 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr12_+_53835383 | 0.00 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr2_+_234104079 | 0.00 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr10_+_124320156 | 0.00 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chrX_+_56590002 | 0.00 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr12_+_53835539 | 0.00 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr2_+_204732487 | 0.00 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr18_-_72920372 | 0.00 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr16_-_11492366 | 0.00 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr12_+_53835508 | 0.00 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr10_+_124320195 | 0.00 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr6_+_160211481 | 0.00 |
ENST00000367034.4
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chr3_+_129159039 | 0.00 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr3_-_129158676 | 0.00 |
ENST00000393278.2
|
MBD4
|
methyl-CpG binding domain protein 4 |
| chr11_+_68228186 | 0.00 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_-_115323245 | 0.00 |
ENST00000060969.5
ENST00000369528.5 |
SIKE1
|
suppressor of IKBKE 1 |
| chr17_-_29151794 | 0.00 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr14_+_64854958 | 0.00 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr2_+_44396000 | 0.00 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |