Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PAX8
Z-value: 0.80
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg19_v2_chr2_-_113993020_113993097 | -0.40 | 6.0e-01 | Click! |
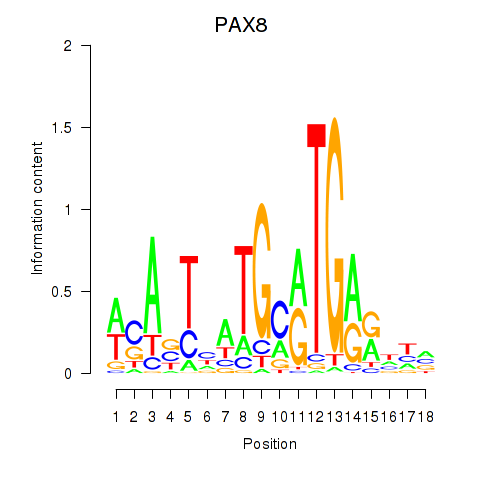
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_24126350 | 1.24 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr11_-_9482010 | 0.78 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr20_-_36794938 | 0.69 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr6_-_27858570 | 0.67 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr11_+_59705928 | 0.59 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr21_+_45287112 | 0.52 |
ENST00000448287.1
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr11_-_118272610 | 0.41 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr19_+_34891252 | 0.39 |
ENST00000606020.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr7_+_140396756 | 0.37 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr22_-_44258360 | 0.35 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr3_-_107941209 | 0.35 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr7_+_140396946 | 0.34 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr1_+_218683438 | 0.32 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr2_+_90198535 | 0.30 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr1_+_180165672 | 0.29 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_+_131549610 | 0.27 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr19_+_9361606 | 0.26 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr7_+_140396465 | 0.25 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr6_-_44233361 | 0.25 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr11_+_63742050 | 0.25 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr19_-_55791431 | 0.24 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr10_-_128110441 | 0.24 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr19_+_50169216 | 0.23 |
ENST00000594157.1
ENST00000600947.1 ENST00000598306.1 |
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr21_-_44582378 | 0.22 |
ENST00000433840.1
|
AP001631.10
|
AP001631.10 |
| chr14_-_54425475 | 0.22 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr19_+_50169081 | 0.22 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr11_-_62609281 | 0.21 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr8_-_66474884 | 0.21 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr2_+_90192768 | 0.21 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr19_-_55791540 | 0.21 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr9_-_21228221 | 0.20 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr1_-_182573514 | 0.20 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr17_-_42100474 | 0.20 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr10_+_35484793 | 0.19 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr1_+_16083154 | 0.19 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_-_73505961 | 0.19 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chr9_-_116837249 | 0.19 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr1_-_153514241 | 0.19 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr9_-_128246769 | 0.18 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr17_+_7461613 | 0.18 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_171283331 | 0.17 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr12_+_72058130 | 0.17 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr1_+_161494036 | 0.17 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr2_+_173724771 | 0.16 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_16083098 | 0.15 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_-_87350970 | 0.15 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr3_-_112329110 | 0.15 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr19_-_36019123 | 0.15 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr9_+_124048864 | 0.15 |
ENST00000545652.1
|
GSN
|
gelsolin |
| chr3_-_137851220 | 0.14 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr1_-_8585945 | 0.14 |
ENST00000377464.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr11_-_62457371 | 0.14 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr2_-_128615517 | 0.14 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr9_-_92020841 | 0.13 |
ENST00000433650.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr13_+_31309645 | 0.13 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr19_+_41856816 | 0.13 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr1_+_16083123 | 0.13 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr18_-_58040000 | 0.13 |
ENST00000299766.3
|
MC4R
|
melanocortin 4 receptor |
| chr1_-_20755140 | 0.13 |
ENST00000418743.1
ENST00000426428.1 |
RP4-749H3.1
|
long intergenic non-protein coding RNA 1141 |
| chr19_+_35939154 | 0.13 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr20_+_4152356 | 0.11 |
ENST00000379460.2
|
SMOX
|
spermine oxidase |
| chr1_+_154300217 | 0.11 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr19_+_17337473 | 0.11 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr5_-_140027357 | 0.11 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr12_-_123201337 | 0.11 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr7_-_144533074 | 0.11 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr11_+_67055986 | 0.11 |
ENST00000447274.2
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr17_-_73267304 | 0.11 |
ENST00000579297.1
ENST00000580571.1 |
MIF4GD
|
MIF4G domain containing |
| chr20_-_3762087 | 0.10 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr16_-_18911366 | 0.10 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_+_161993465 | 0.10 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_73266616 | 0.10 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr7_+_90012986 | 0.10 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr4_-_23735183 | 0.10 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr2_+_38177575 | 0.10 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr19_-_51487282 | 0.10 |
ENST00000595820.1
ENST00000597707.1 ENST00000336317.4 |
KLK7
|
kallikrein-related peptidase 7 |
| chr8_+_124429006 | 0.10 |
ENST00000522194.1
ENST00000523356.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr19_+_40476912 | 0.09 |
ENST00000157812.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr16_+_318638 | 0.09 |
ENST00000412541.1
ENST00000435035.1 |
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr8_+_21915368 | 0.09 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr12_-_110883346 | 0.09 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr11_-_129062093 | 0.09 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr14_-_91710852 | 0.09 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr5_-_176943917 | 0.09 |
ENST00000330503.7
|
DDX41
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
| chr16_-_30773372 | 0.09 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr18_-_21017817 | 0.09 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr4_+_72204755 | 0.09 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr16_+_2083265 | 0.09 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr1_-_25256368 | 0.09 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr16_+_57496299 | 0.09 |
ENST00000219252.5
|
POLR2C
|
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
| chr1_-_71513471 | 0.09 |
ENST00000370931.3
ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr4_-_75695366 | 0.08 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr19_+_782755 | 0.08 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr15_+_45028520 | 0.08 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr1_+_160321120 | 0.08 |
ENST00000424754.1
|
NCSTN
|
nicastrin |
| chr19_+_54704610 | 0.08 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr6_+_167704838 | 0.08 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr18_-_53068782 | 0.08 |
ENST00000569012.1
|
TCF4
|
transcription factor 4 |
| chr19_+_54704718 | 0.08 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr17_-_34257771 | 0.08 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr17_+_48624450 | 0.08 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr22_-_20138302 | 0.08 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr6_-_31628512 | 0.08 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr17_+_1933404 | 0.08 |
ENST00000263083.6
ENST00000571418.1 |
DPH1
|
diphthamide biosynthesis 1 |
| chr13_+_27844464 | 0.08 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr2_-_106013400 | 0.08 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr17_-_19648916 | 0.08 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr12_+_128399965 | 0.08 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr7_-_76255444 | 0.08 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr12_-_6960407 | 0.08 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr11_-_2170786 | 0.08 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr5_+_140235469 | 0.08 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr17_+_7461849 | 0.08 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_-_7812446 | 0.07 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr19_-_50168962 | 0.07 |
ENST00000599223.1
ENST00000593922.1 ENST00000600022.1 ENST00000596765.1 ENST00000599144.1 ENST00000596822.1 ENST00000598108.1 ENST00000601373.1 ENST00000595034.1 ENST00000601291.1 |
IRF3
|
interferon regulatory factor 3 |
| chr3_-_150264272 | 0.07 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr17_+_7462103 | 0.07 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_-_55791563 | 0.07 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr6_-_134639180 | 0.07 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr20_-_1373682 | 0.07 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr19_+_17865011 | 0.07 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr1_+_155006300 | 0.07 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr10_+_12391685 | 0.07 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr4_-_10686475 | 0.07 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr12_+_128399917 | 0.07 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr6_+_161123270 | 0.07 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr16_-_31519691 | 0.07 |
ENST00000567994.1
ENST00000430477.2 ENST00000570164.1 ENST00000327237.2 |
C16orf58
|
chromosome 16 open reading frame 58 |
| chr19_+_52901094 | 0.06 |
ENST00000391788.2
ENST00000436397.1 ENST00000391787.2 ENST00000360465.3 ENST00000494167.2 ENST00000493272.1 |
ZNF528
|
zinc finger protein 528 |
| chrX_-_84363974 | 0.06 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr3_-_49158218 | 0.06 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr12_-_129308487 | 0.06 |
ENST00000266771.5
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr6_+_167704798 | 0.06 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr18_-_29264467 | 0.06 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr3_-_178969403 | 0.06 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr17_-_67057114 | 0.06 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr6_+_30844192 | 0.06 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_+_122516626 | 0.06 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr17_+_6918093 | 0.06 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr2_+_85661918 | 0.06 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr3_-_49158312 | 0.06 |
ENST00000398892.3
ENST00000453664.1 ENST00000398888.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr20_+_58571419 | 0.06 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr6_+_135502408 | 0.06 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr5_+_69321074 | 0.06 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr19_-_7812397 | 0.05 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr7_+_95115210 | 0.05 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr2_-_37068530 | 0.05 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr1_+_367640 | 0.05 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr17_-_73267214 | 0.05 |
ENST00000580717.1
ENST00000577542.1 ENST00000579612.1 ENST00000245551.5 |
MIF4GD
|
MIF4G domain containing |
| chr9_+_42704004 | 0.05 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chr11_+_65292538 | 0.05 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr17_-_67057203 | 0.05 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr2_+_219524473 | 0.05 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr8_-_124741451 | 0.05 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr22_-_31324215 | 0.05 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr8_-_121825088 | 0.05 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr7_-_112430647 | 0.05 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr17_-_80017856 | 0.05 |
ENST00000577574.1
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr7_-_128001658 | 0.05 |
ENST00000489835.2
ENST00000464607.1 ENST00000489517.1 ENST00000446477.2 ENST00000535159.1 ENST00000435512.1 ENST00000495931.1 |
PRRT4
|
proline-rich transmembrane protein 4 |
| chr21_-_43816052 | 0.05 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr19_-_35992780 | 0.05 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr19_+_42041860 | 0.05 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr13_-_103426112 | 0.04 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr12_-_6961050 | 0.04 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr16_+_75690093 | 0.04 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr17_-_8301132 | 0.04 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chrX_-_23926004 | 0.04 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr17_-_3499125 | 0.04 |
ENST00000399759.3
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr17_-_73267281 | 0.04 |
ENST00000578305.1
ENST00000325102.8 |
MIF4GD
|
MIF4G domain containing |
| chr18_-_53068940 | 0.04 |
ENST00000562638.1
|
TCF4
|
transcription factor 4 |
| chr17_-_60883993 | 0.04 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr4_+_118955500 | 0.04 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr5_+_148521136 | 0.04 |
ENST00000506113.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr4_+_71600144 | 0.04 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr19_+_35417939 | 0.04 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr10_-_5638048 | 0.03 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chrM_+_4431 | 0.03 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr4_-_69536346 | 0.03 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr11_+_20044600 | 0.03 |
ENST00000311043.8
|
NAV2
|
neuron navigator 2 |
| chr3_+_186435137 | 0.03 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr5_-_156390230 | 0.03 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr4_-_128887069 | 0.03 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr4_+_69681710 | 0.03 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr2_-_106013207 | 0.03 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr1_+_41448820 | 0.03 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr17_-_74023291 | 0.03 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr6_+_135502466 | 0.03 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_+_20044096 | 0.03 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr8_+_124428959 | 0.03 |
ENST00000287387.2
ENST00000523984.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr17_+_5031687 | 0.03 |
ENST00000250066.6
ENST00000304328.5 |
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene) |
| chr12_-_11287243 | 0.03 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr3_-_8811288 | 0.03 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr2_+_171036635 | 0.03 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr6_+_147527103 | 0.02 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr22_-_50523688 | 0.02 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr10_+_70847852 | 0.02 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr15_-_60683326 | 0.02 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr11_-_58343319 | 0.02 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr7_-_47520869 | 0.02 |
ENST00000415929.1
|
TNS3
|
tensin 3 |
| chr11_+_60609537 | 0.02 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr6_+_31126291 | 0.02 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) transcription from RNA polymerase I promoter(GO:0006360) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.2 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.4 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0002778 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.0 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0070560 | calcium-mediated signaling using extracellular calcium source(GO:0035585) protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |