Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
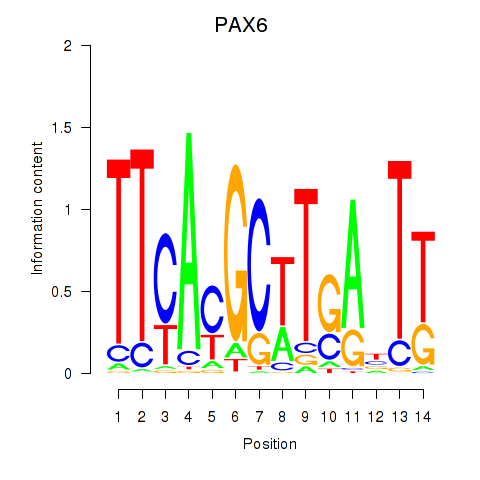
Results for PAX6
Z-value: 0.79
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.16 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX6 | hg19_v2_chr11_-_31839488_31839515 | 0.83 | 1.7e-01 | Click! |
Activity profile of PAX6 motif
Sorted Z-values of PAX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35820064 | 0.39 |
ENST00000341773.6
ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22
|
CD22 molecule |
| chr1_-_43282945 | 0.33 |
ENST00000537227.1
|
CCDC23
|
coiled-coil domain containing 23 |
| chr2_-_202298268 | 0.32 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr16_-_80926457 | 0.30 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr5_-_119669160 | 0.30 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr4_+_70916119 | 0.30 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr8_+_67104323 | 0.29 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr3_-_196910477 | 0.27 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_-_204183071 | 0.27 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr3_+_108015382 | 0.26 |
ENST00000463019.3
ENST00000491820.1 ENST00000467562.1 ENST00000482430.1 ENST00000462629.1 |
HHLA2
|
HERV-H LTR-associating 2 |
| chr12_-_70093235 | 0.24 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr19_+_42037433 | 0.23 |
ENST00000599316.1
ENST00000599770.1 |
AC006129.1
|
AC006129.1 |
| chr17_-_58499766 | 0.23 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr3_+_172034218 | 0.23 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr2_-_176867534 | 0.22 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr6_-_42162654 | 0.21 |
ENST00000230361.3
|
GUCA1B
|
guanylate cyclase activator 1B (retina) |
| chr11_+_1295809 | 0.21 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr1_-_156269428 | 0.19 |
ENST00000339922.3
|
VHLL
|
von Hippel-Lindau tumor suppressor-like |
| chr3_-_195310802 | 0.19 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr16_-_71264558 | 0.18 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
| chr12_-_3982548 | 0.18 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr1_-_55089191 | 0.17 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr12_-_70093111 | 0.17 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr6_-_52859046 | 0.17 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_+_109204743 | 0.17 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_171489085 | 0.16 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr2_+_149402989 | 0.16 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr14_+_61995722 | 0.15 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr6_+_26225354 | 0.15 |
ENST00000360408.1
|
HIST1H3E
|
histone cluster 1, H3e |
| chr19_+_3708338 | 0.15 |
ENST00000590545.1
|
TJP3
|
tight junction protein 3 |
| chr3_-_15563229 | 0.15 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr22_-_29949657 | 0.14 |
ENST00000428374.1
|
THOC5
|
THO complex 5 |
| chr15_+_42565393 | 0.14 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr2_+_191334212 | 0.14 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr8_-_95487272 | 0.14 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr7_-_102252589 | 0.14 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr7_-_91808845 | 0.13 |
ENST00000343318.5
|
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr17_-_39041479 | 0.13 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr3_-_119379719 | 0.12 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr10_+_13628933 | 0.12 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr1_+_203765437 | 0.12 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr7_+_65552756 | 0.12 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr5_-_68339648 | 0.11 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr7_-_84121858 | 0.11 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_-_118765081 | 0.11 |
ENST00000392903.2
ENST00000355371.4 |
KIAA1598
|
KIAA1598 |
| chr14_+_21569245 | 0.11 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr8_+_41347915 | 0.11 |
ENST00000518270.1
ENST00000520817.1 |
GOLGA7
|
golgin A7 |
| chr2_-_153573965 | 0.11 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr12_+_83080724 | 0.11 |
ENST00000548305.1
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr2_-_161056762 | 0.11 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr13_+_25875785 | 0.11 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr16_-_51185172 | 0.10 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr10_-_75226166 | 0.10 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr2_+_114195268 | 0.10 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr6_-_131291572 | 0.10 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr10_-_27389320 | 0.10 |
ENST00000436985.2
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr5_+_322685 | 0.10 |
ENST00000510400.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr20_-_62178857 | 0.10 |
ENST00000217188.1
|
SRMS
|
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites |
| chr8_-_95487331 | 0.09 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr10_+_12110963 | 0.09 |
ENST00000263035.4
ENST00000437298.1 |
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr11_-_11374904 | 0.09 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr4_+_40751914 | 0.09 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chr5_-_98262240 | 0.09 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr5_-_173173171 | 0.09 |
ENST00000517733.1
ENST00000518902.1 |
CTB-43E15.3
|
CTB-43E15.3 |
| chr4_-_65275162 | 0.09 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr7_+_102292796 | 0.09 |
ENST00000436228.2
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr4_-_129491686 | 0.09 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr15_-_78358465 | 0.09 |
ENST00000435468.1
|
TBC1D2B
|
TBC1 domain family, member 2B |
| chr8_-_54934708 | 0.09 |
ENST00000520534.1
ENST00000518784.1 ENST00000522635.1 |
TCEA1
|
transcription elongation factor A (SII), 1 |
| chr3_-_196910721 | 0.09 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_-_27389392 | 0.09 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr1_+_104068562 | 0.09 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr1_-_36906474 | 0.08 |
ENST00000433045.2
|
OSCP1
|
organic solute carrier partner 1 |
| chr10_-_118764862 | 0.08 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr4_-_798689 | 0.08 |
ENST00000504062.1
|
CPLX1
|
complexin 1 |
| chr8_+_66955648 | 0.08 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr1_-_89458287 | 0.08 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr8_+_121457642 | 0.08 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr8_-_27469383 | 0.08 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr7_-_80551671 | 0.07 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_+_79115503 | 0.07 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr17_-_77023683 | 0.07 |
ENST00000581579.1
|
C1QTNF1-AS1
|
C1QTNF1 antisense RNA 1 |
| chr6_-_88299678 | 0.07 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr12_+_56915713 | 0.07 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chrX_+_10126488 | 0.07 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr1_+_203651937 | 0.07 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr6_-_31846744 | 0.07 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr22_-_36220420 | 0.07 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_122247879 | 0.07 |
ENST00000545861.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr12_+_49740700 | 0.07 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr3_-_15540055 | 0.07 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr21_+_40752378 | 0.06 |
ENST00000398753.1
ENST00000442773.1 |
WRB
|
tryptophan rich basic protein |
| chr1_+_207925391 | 0.06 |
ENST00000358170.2
ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46
|
CD46 molecule, complement regulatory protein |
| chr2_-_162931052 | 0.06 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr12_-_10007448 | 0.06 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr18_+_61420169 | 0.06 |
ENST00000425392.1
ENST00000336429.2 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr6_+_125474992 | 0.06 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr10_-_17243579 | 0.06 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr9_+_127023704 | 0.06 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr13_+_96085847 | 0.06 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr12_-_106697974 | 0.06 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr1_-_89458415 | 0.06 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr5_+_119799927 | 0.06 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr1_+_223354486 | 0.06 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr10_-_13523073 | 0.06 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr1_-_43282906 | 0.06 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr18_-_21242729 | 0.06 |
ENST00000585908.2
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr17_-_77023723 | 0.06 |
ENST00000577521.1
|
C1QTNF1-AS1
|
C1QTNF1 antisense RNA 1 |
| chr3_-_195997410 | 0.06 |
ENST00000419333.1
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr18_-_21242774 | 0.06 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr19_+_36266433 | 0.06 |
ENST00000314737.5
|
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr2_+_196521903 | 0.06 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_+_196521845 | 0.06 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr20_-_1472029 | 0.05 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr7_+_142919130 | 0.05 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr6_+_170151708 | 0.05 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr10_+_102222798 | 0.05 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr6_-_43027105 | 0.05 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr1_-_184006829 | 0.05 |
ENST00000361927.4
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr3_-_119182506 | 0.05 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr6_+_125474939 | 0.05 |
ENST00000527711.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr20_+_62887081 | 0.05 |
ENST00000369758.4
ENST00000299468.7 ENST00000609372.1 ENST00000610196.1 ENST00000308824.6 |
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr9_-_13279563 | 0.05 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr7_-_97501706 | 0.05 |
ENST00000455086.1
ENST00000453600.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr2_+_196521458 | 0.05 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr8_+_18067602 | 0.05 |
ENST00000307719.4
ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr7_+_133615169 | 0.05 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr2_+_29320571 | 0.05 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr21_-_35014027 | 0.05 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr17_-_76824975 | 0.05 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr2_+_192109911 | 0.05 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr9_-_116861337 | 0.05 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr6_-_131321863 | 0.05 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr13_+_24844857 | 0.05 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr1_+_158901329 | 0.05 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr6_+_88299833 | 0.05 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr2_-_136743169 | 0.05 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr12_+_111284764 | 0.04 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr2_+_192110199 | 0.04 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr4_+_113970772 | 0.04 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr7_-_97501733 | 0.04 |
ENST00000444334.1
ENST00000422745.1 ENST00000394308.3 ENST00000451771.1 ENST00000175506.4 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr13_+_25875662 | 0.04 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr11_-_57298187 | 0.04 |
ENST00000525158.1
ENST00000257245.4 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr16_-_47493041 | 0.04 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr8_-_19615382 | 0.04 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_-_88141615 | 0.04 |
ENST00000545252.1
ENST00000425278.2 ENST00000498875.2 |
KLHL8
|
kelch-like family member 8 |
| chr12_-_122107549 | 0.04 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr6_-_26033796 | 0.04 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr5_-_159546396 | 0.04 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr5_+_94982558 | 0.04 |
ENST00000311364.4
ENST00000458310.1 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr1_-_8586084 | 0.04 |
ENST00000464972.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr1_+_17248418 | 0.04 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr13_+_24844979 | 0.04 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr11_+_57105991 | 0.04 |
ENST00000263314.2
|
P2RX3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr15_+_52043758 | 0.04 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr19_-_37701386 | 0.04 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chr5_+_140602904 | 0.04 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr10_+_13628921 | 0.04 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr16_-_18937726 | 0.04 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr14_+_56127960 | 0.04 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr8_+_142402089 | 0.04 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr12_+_83080659 | 0.04 |
ENST00000321196.3
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr17_-_8661860 | 0.04 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr1_-_165738072 | 0.04 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr3_+_15045419 | 0.04 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr15_-_23034322 | 0.04 |
ENST00000539711.2
ENST00000560039.1 ENST00000398013.3 ENST00000337451.3 ENST00000359727.4 ENST00000398014.2 |
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chr6_-_10412600 | 0.03 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr9_+_129097520 | 0.03 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr5_-_58882219 | 0.03 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr14_+_24099318 | 0.03 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr3_-_142297668 | 0.03 |
ENST00000350721.4
ENST00000383101.3 |
ATR
|
ataxia telangiectasia and Rad3 related |
| chr6_-_134373732 | 0.03 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr1_-_85725316 | 0.03 |
ENST00000344356.5
ENST00000471115.1 |
C1orf52
|
chromosome 1 open reading frame 52 |
| chrX_-_138914394 | 0.03 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr17_+_36283971 | 0.03 |
ENST00000327454.6
ENST00000378174.5 |
TBC1D3F
|
TBC1 domain family, member 3F |
| chr19_+_3708376 | 0.03 |
ENST00000539908.2
|
TJP3
|
tight junction protein 3 |
| chr4_+_130017268 | 0.03 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr2_-_220110111 | 0.03 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr4_-_187517928 | 0.03 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr21_-_18985230 | 0.03 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr8_-_62602327 | 0.03 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr3_-_119182523 | 0.03 |
ENST00000319172.5
|
TMEM39A
|
transmembrane protein 39A |
| chr11_-_117688216 | 0.03 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr11_+_33563821 | 0.03 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr10_+_114135952 | 0.03 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr5_+_322733 | 0.03 |
ENST00000515206.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr1_-_229644034 | 0.03 |
ENST00000366678.3
ENST00000261396.3 ENST00000537506.1 |
NUP133
|
nucleoporin 133kDa |
| chr2_-_74776586 | 0.03 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr11_+_65554493 | 0.03 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr16_-_71518504 | 0.03 |
ENST00000565100.2
|
ZNF19
|
zinc finger protein 19 |
| chr4_-_69215467 | 0.03 |
ENST00000579690.1
|
YTHDC1
|
YTH domain containing 1 |
| chr11_-_102401469 | 0.02 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_-_110933611 | 0.02 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr15_+_52155001 | 0.02 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr12_-_56367101 | 0.02 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr12_-_56693758 | 0.02 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr16_-_3767506 | 0.02 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr11_+_33563618 | 0.02 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr17_-_73389737 | 0.02 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr6_-_33041378 | 0.02 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.2 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.3 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.0 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0036343 | negative regulation of extracellular matrix disassembly(GO:0010716) psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.0 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |