Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
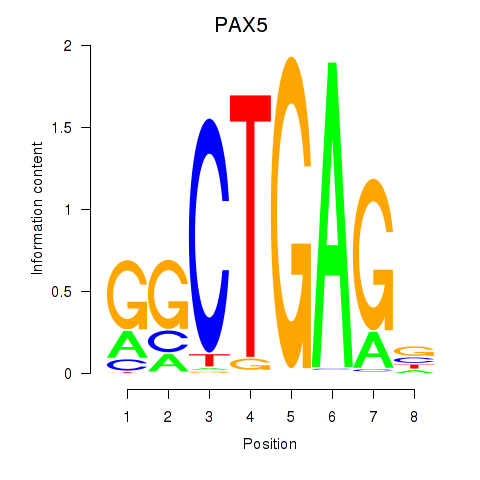
Results for PAX5
Z-value: 0.26
Transcription factors associated with PAX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX5
|
ENSG00000196092.8 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX5 | hg19_v2_chr9_-_37034028_37034157 | -0.13 | 8.7e-01 | Click! |
Activity profile of PAX5 motif
Sorted Z-values of PAX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_7281469 | 0.14 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr19_+_1261106 | 0.11 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr4_+_76649753 | 0.11 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr11_+_117198801 | 0.10 |
ENST00000527609.1
ENST00000533570.1 |
CEP164
|
centrosomal protein 164kDa |
| chr19_+_49838653 | 0.10 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr19_-_54618650 | 0.10 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr17_-_58469591 | 0.10 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr22_+_20905422 | 0.10 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr15_+_101417919 | 0.09 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr22_-_37882395 | 0.09 |
ENST00000416983.3
ENST00000424765.2 ENST00000356998.3 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_-_14322319 | 0.09 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chrX_+_102024075 | 0.09 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr16_-_11723066 | 0.09 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr9_+_131445703 | 0.09 |
ENST00000454747.1
|
SET
|
SET nuclear oncogene |
| chr10_-_112255945 | 0.08 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr1_-_1208851 | 0.08 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr3_+_187930429 | 0.08 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr10_-_75532373 | 0.08 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr1_-_52520828 | 0.08 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr9_+_130159433 | 0.08 |
ENST00000451404.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr6_-_34524093 | 0.08 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr14_-_23288930 | 0.08 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr8_+_97657531 | 0.08 |
ENST00000519900.1
ENST00000517742.1 |
CPQ
|
carboxypeptidase Q |
| chr14_-_23791484 | 0.08 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr17_-_27277615 | 0.08 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr2_-_3595547 | 0.07 |
ENST00000438485.1
|
RP13-512J5.1
|
Uncharacterized protein |
| chr2_-_11272234 | 0.07 |
ENST00000590207.1
ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1
|
AC062028.1 |
| chr20_+_34679725 | 0.07 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr15_+_91418918 | 0.07 |
ENST00000560824.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_+_120839005 | 0.07 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr3_-_142720267 | 0.07 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr8_+_144373550 | 0.07 |
ENST00000330143.3
ENST00000521537.1 ENST00000518432.1 ENST00000520333.1 |
ZNF696
|
zinc finger protein 696 |
| chr17_+_4692230 | 0.07 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr1_+_249132512 | 0.07 |
ENST00000505503.1
|
ZNF672
|
zinc finger protein 672 |
| chr21_+_44866471 | 0.07 |
ENST00000448049.1
|
LINC00319
|
long intergenic non-protein coding RNA 319 |
| chr19_-_46272462 | 0.07 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr1_+_53793885 | 0.06 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr6_-_85473073 | 0.06 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr19_-_56048456 | 0.06 |
ENST00000413299.1
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr12_+_56114151 | 0.06 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr18_-_2982869 | 0.06 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr9_+_130159471 | 0.06 |
ENST00000419917.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr11_-_69490135 | 0.06 |
ENST00000542341.1
|
ORAOV1
|
oral cancer overexpressed 1 |
| chr16_-_19725899 | 0.06 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr16_-_1429010 | 0.06 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr9_+_35673853 | 0.06 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr3_+_49840685 | 0.06 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr5_-_138730817 | 0.06 |
ENST00000434752.2
|
PROB1
|
proline-rich basic protein 1 |
| chr16_-_11692320 | 0.06 |
ENST00000571627.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr18_+_12658185 | 0.06 |
ENST00000400514.2
|
AP005482.1
|
Uncharacterized protein |
| chr19_-_49828438 | 0.06 |
ENST00000454748.3
ENST00000598828.1 ENST00000335875.4 |
SLC6A16
|
solute carrier family 6, member 16 |
| chr14_-_91294472 | 0.06 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chr3_-_52739670 | 0.06 |
ENST00000497953.1
|
GLT8D1
|
glycosyltransferase 8 domain containing 1 |
| chr16_-_70712229 | 0.06 |
ENST00000562883.2
|
MTSS1L
|
metastasis suppressor 1-like |
| chr1_+_197871854 | 0.06 |
ENST00000436652.1
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr17_-_64216748 | 0.06 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_-_987164 | 0.06 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr9_-_138393580 | 0.06 |
ENST00000371791.1
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr5_+_56205878 | 0.06 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr2_-_74730430 | 0.06 |
ENST00000460508.3
|
LBX2
|
ladybird homeobox 2 |
| chr1_-_78444776 | 0.06 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr12_-_52604607 | 0.06 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr3_+_187930491 | 0.06 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr9_-_33402506 | 0.06 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr1_+_901847 | 0.06 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr22_+_22676808 | 0.06 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_+_128177458 | 0.06 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr5_+_76012009 | 0.06 |
ENST00000505600.1
|
F2R
|
coagulation factor II (thrombin) receptor |
| chr19_-_53193731 | 0.05 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr3_-_48632593 | 0.05 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr3_-_135915146 | 0.05 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr1_-_78444738 | 0.05 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr17_+_4846101 | 0.05 |
ENST00000576965.1
|
RNF167
|
ring finger protein 167 |
| chr19_+_46009837 | 0.05 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr7_-_105926058 | 0.05 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr1_+_27113963 | 0.05 |
ENST00000430292.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr14_+_94577074 | 0.05 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr14_-_62162541 | 0.05 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr11_+_62475130 | 0.05 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr16_-_67217844 | 0.05 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr7_+_7222157 | 0.05 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr17_+_80193644 | 0.05 |
ENST00000582946.1
|
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr6_-_34524049 | 0.05 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr4_+_169633310 | 0.05 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr15_-_52971544 | 0.05 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr2_-_97509749 | 0.05 |
ENST00000331001.2
ENST00000318357.4 |
ANKRD23
|
ankyrin repeat domain 23 |
| chr3_-_120400960 | 0.05 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr8_-_94753202 | 0.05 |
ENST00000521947.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr14_-_23285069 | 0.05 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_+_71295766 | 0.05 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr11_+_119038897 | 0.05 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr17_-_4900873 | 0.05 |
ENST00000355025.3
ENST00000575780.1 ENST00000396829.2 |
INCA1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr9_+_140145713 | 0.05 |
ENST00000388931.3
ENST00000412566.1 |
C9orf173
|
chromosome 9 open reading frame 173 |
| chr14_-_75530693 | 0.05 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr9_+_130159593 | 0.05 |
ENST00000419132.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr1_+_2121228 | 0.05 |
ENST00000597060.1
|
AL590822.2
|
Uncharacterized protein; cDNA FLJ36608 fis, clone TRACH2015824 |
| chr1_-_16556038 | 0.05 |
ENST00000375605.2
|
C1orf134
|
chromosome 1 open reading frame 134 |
| chr20_+_19738792 | 0.05 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr4_-_75024085 | 0.05 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr19_+_1954632 | 0.05 |
ENST00000589350.1
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr19_-_47735918 | 0.05 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr19_+_4343524 | 0.05 |
ENST00000262966.8
ENST00000359935.4 ENST00000599840.1 |
MPND
|
MPN domain containing |
| chr12_+_123949053 | 0.05 |
ENST00000350887.5
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr1_+_206138457 | 0.05 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr19_+_1000418 | 0.05 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr14_-_75079294 | 0.04 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr15_+_76030311 | 0.04 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr2_-_74648702 | 0.04 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr17_+_46985823 | 0.04 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr11_+_45168182 | 0.04 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr14_+_52327350 | 0.04 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr19_-_51872233 | 0.04 |
ENST00000601435.1
ENST00000291715.1 |
CLDND2
|
claudin domain containing 2 |
| chrX_-_77150911 | 0.04 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr2_+_219221573 | 0.04 |
ENST00000289388.3
|
C2orf62
|
chromosome 2 open reading frame 62 |
| chr19_+_18723660 | 0.04 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chrX_-_70288234 | 0.04 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr7_+_150748288 | 0.04 |
ENST00000490540.1
|
ASIC3
|
acid-sensing (proton-gated) ion channel 3 |
| chr15_+_41057818 | 0.04 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr1_+_20878932 | 0.04 |
ENST00000332947.4
|
FAM43B
|
family with sequence similarity 43, member B |
| chr5_-_138780159 | 0.04 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr16_-_28223229 | 0.04 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr7_+_100728720 | 0.04 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr15_+_44829334 | 0.04 |
ENST00000535391.1
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J |
| chr3_-_50605077 | 0.04 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr10_-_75385711 | 0.04 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr7_-_150038704 | 0.04 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr2_+_136499287 | 0.04 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr15_-_40574787 | 0.04 |
ENST00000434396.1
|
ANKRD63
|
ankyrin repeat domain 63 |
| chr1_-_8939265 | 0.04 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr19_+_38893751 | 0.04 |
ENST00000588262.1
ENST00000252530.5 ENST00000343358.7 |
FAM98C
|
family with sequence similarity 98, member C |
| chr6_+_53659877 | 0.04 |
ENST00000370882.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr10_-_18948156 | 0.04 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr19_-_6481776 | 0.04 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr2_+_233404429 | 0.04 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr8_-_11324273 | 0.04 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr6_+_29691056 | 0.04 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr17_+_18163848 | 0.04 |
ENST00000323019.4
ENST00000578174.1 ENST00000395704.4 ENST00000395703.4 ENST00000578621.1 ENST00000579341.1 |
MIEF2
|
mitochondrial elongation factor 2 |
| chr1_-_156571254 | 0.04 |
ENST00000438976.2
ENST00000334588.7 ENST00000368232.4 ENST00000415314.2 |
GPATCH4
|
G patch domain containing 4 |
| chr8_+_22423479 | 0.04 |
ENST00000522721.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_-_143913143 | 0.04 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr17_-_41322332 | 0.04 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr6_-_107235331 | 0.04 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr19_+_46144884 | 0.04 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr11_+_844406 | 0.04 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr16_+_30671223 | 0.04 |
ENST00000568722.1
|
FBRS
|
fibrosin |
| chr19_-_55895966 | 0.04 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238 |
| chr8_-_142377367 | 0.04 |
ENST00000377741.3
|
GPR20
|
G protein-coupled receptor 20 |
| chr2_+_74881432 | 0.04 |
ENST00000453930.1
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr22_-_38966123 | 0.04 |
ENST00000439567.1
|
DMC1
|
DNA meiotic recombinase 1 |
| chr12_+_49740700 | 0.04 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr19_-_10213335 | 0.04 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr3_-_9811674 | 0.04 |
ENST00000411972.1
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr5_-_139944196 | 0.04 |
ENST00000357560.4
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr3_-_129147432 | 0.04 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr11_-_69490073 | 0.04 |
ENST00000535657.1
ENST00000539414.1 ENST00000536870.1 ENST00000538554.2 ENST00000279147.4 |
ORAOV1
|
oral cancer overexpressed 1 |
| chr3_-_50605150 | 0.04 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr8_+_22428457 | 0.04 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr14_-_50999190 | 0.04 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_+_60758814 | 0.04 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr11_-_128775930 | 0.04 |
ENST00000524878.1
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chr19_-_58514129 | 0.04 |
ENST00000552184.1
ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606
|
zinc finger protein 606 |
| chr20_-_1165117 | 0.04 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr11_-_8739383 | 0.04 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr19_+_38893809 | 0.04 |
ENST00000589408.1
|
FAM98C
|
family with sequence similarity 98, member C |
| chr12_+_123942188 | 0.04 |
ENST00000526639.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr2_+_65663812 | 0.04 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr20_-_32580924 | 0.04 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr19_+_54606145 | 0.04 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr6_+_30848740 | 0.04 |
ENST00000505534.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_-_49837254 | 0.04 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr12_+_6930703 | 0.04 |
ENST00000311268.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr8_-_94752946 | 0.04 |
ENST00000519109.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr22_-_30198075 | 0.04 |
ENST00000411532.1
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr1_-_1763721 | 0.04 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr8_+_72740402 | 0.04 |
ENST00000521467.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr2_+_74881398 | 0.04 |
ENST00000339773.5
ENST00000434486.1 |
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr7_+_44040488 | 0.04 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr7_+_70229899 | 0.04 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr10_+_43932553 | 0.04 |
ENST00000456416.1
ENST00000437590.2 ENST00000451167.1 |
ZNF487
|
zinc finger protein 487 |
| chr14_+_24779376 | 0.04 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr13_+_110958124 | 0.04 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr11_+_695787 | 0.04 |
ENST00000526170.1
ENST00000488769.1 |
TMEM80
|
transmembrane protein 80 |
| chr22_-_29663954 | 0.04 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chrX_+_48660565 | 0.04 |
ENST00000413163.2
ENST00000441703.1 ENST00000426196.1 |
HDAC6
|
histone deacetylase 6 |
| chr11_+_64052294 | 0.04 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr19_-_49339080 | 0.04 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_-_43690642 | 0.04 |
ENST00000407356.1
ENST00000407568.1 ENST00000404580.1 ENST00000599812.1 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chr1_+_109642799 | 0.04 |
ENST00000602755.1
|
SCARNA2
|
small Cajal body-specific RNA 2 |
| chrX_-_83757399 | 0.04 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chrX_-_70331298 | 0.04 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr16_+_89979826 | 0.04 |
ENST00000555427.1
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr12_-_53473136 | 0.04 |
ENST00000547837.1
ENST00000301463.4 |
SPRYD3
|
SPRY domain containing 3 |
| chr5_-_126409159 | 0.04 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr17_-_15165825 | 0.04 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr22_-_29663690 | 0.04 |
ENST00000406335.1
|
RHBDD3
|
rhomboid domain containing 3 |
| chr14_+_78227105 | 0.04 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr4_-_987217 | 0.04 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr14_+_35514323 | 0.04 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr3_-_196910477 | 0.04 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_-_30199516 | 0.04 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.0 | GO:0019732 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.0 | 0.0 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.0 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.0 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.0 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epinephrine secretion(GO:0048242) |
| 0.0 | 0.0 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) trans-synaptic signalling, modulating synaptic transmission(GO:0099550) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.0 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.0 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0061227 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.0 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.0 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.0 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |