Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PAX4
Z-value: 0.44
Transcription factors associated with PAX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX4
|
ENSG00000106331.10 | paired box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX4 | hg19_v2_chr7_-_127255982_127255982 | 0.78 | 2.2e-01 | Click! |
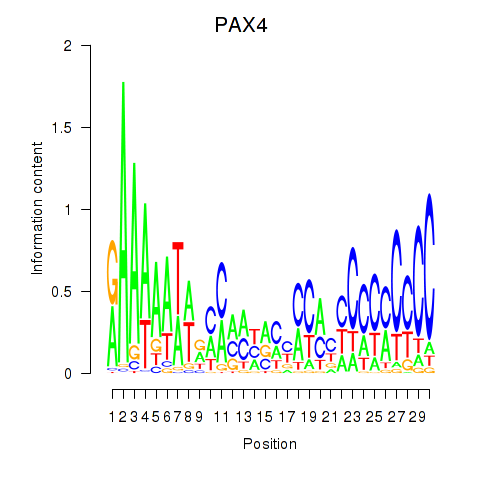
Activity profile of PAX4 motif
Sorted Z-values of PAX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_65314905 | 0.22 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr6_-_31697977 | 0.20 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_-_4859260 | 0.15 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr6_+_31515337 | 0.14 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr19_-_51308175 | 0.14 |
ENST00000345523.4
|
C19orf48
|
chromosome 19 open reading frame 48 |
| chr16_-_3074231 | 0.13 |
ENST00000572355.1
ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr5_-_126409159 | 0.12 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr7_+_72848092 | 0.12 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr1_-_228594490 | 0.12 |
ENST00000366699.3
ENST00000284551.6 |
TRIM11
|
tripartite motif containing 11 |
| chr9_-_139965000 | 0.11 |
ENST00000409687.3
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr1_-_1677358 | 0.11 |
ENST00000355439.2
ENST00000400924.1 ENST00000246421.4 |
SLC35E2
|
solute carrier family 35, member E2 |
| chr17_+_73089382 | 0.11 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr19_-_6767431 | 0.10 |
ENST00000437152.3
ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr3_+_27754397 | 0.10 |
ENST00000606069.1
|
RP11-222K16.2
|
RP11-222K16.2 |
| chr17_+_47448102 | 0.10 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr19_+_9361606 | 0.10 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr16_-_28621353 | 0.09 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_-_123717711 | 0.09 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chrM_+_8366 | 0.09 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr15_+_69373210 | 0.09 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr5_-_172662303 | 0.09 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr22_-_31688431 | 0.09 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr18_+_3448455 | 0.09 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr22_-_31688381 | 0.09 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr7_+_140396465 | 0.08 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr6_-_113754604 | 0.08 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr19_+_14544099 | 0.08 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr19_-_10420459 | 0.08 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr19_-_36391434 | 0.08 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr5_-_172662197 | 0.08 |
ENST00000521848.1
|
NKX2-5
|
NK2 homeobox 5 |
| chr14_+_78227105 | 0.08 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr12_+_56546363 | 0.08 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr4_+_139694701 | 0.08 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr5_-_159766528 | 0.08 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr19_+_35890433 | 0.08 |
ENST00000601669.2
ENST00000441942.2 ENST00000602653.1 ENST00000602796.1 |
AC002511.1
|
AC002511.1 |
| chr10_-_14920599 | 0.07 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr1_+_155146318 | 0.07 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr19_+_2977444 | 0.07 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr17_-_26697304 | 0.07 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr18_-_53255766 | 0.07 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr8_-_145653885 | 0.07 |
ENST00000531032.1
ENST00000292510.4 ENST00000377348.2 ENST00000530790.1 ENST00000533806.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr11_-_62474803 | 0.07 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_+_33895090 | 0.07 |
ENST00000592381.1
|
RP11-1094M14.11
|
RP11-1094M14.11 |
| chr5_-_172662230 | 0.07 |
ENST00000424406.2
|
NKX2-5
|
NK2 homeobox 5 |
| chr4_+_174292058 | 0.06 |
ENST00000296504.3
|
SAP30
|
Sin3A-associated protein, 30kDa |
| chr8_+_21946681 | 0.06 |
ENST00000289921.7
|
FAM160B2
|
family with sequence similarity 160, member B2 |
| chr22_-_39151463 | 0.06 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr2_-_85839146 | 0.06 |
ENST00000306336.5
ENST00000409734.3 |
C2orf68
|
chromosome 2 open reading frame 68 |
| chr6_+_134274354 | 0.06 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr7_+_5468362 | 0.06 |
ENST00000608012.1
|
RP11-1275H24.3
|
RP11-1275H24.3 |
| chr16_-_79804198 | 0.06 |
ENST00000568389.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr7_-_99756293 | 0.06 |
ENST00000316937.3
ENST00000456769.1 |
C7orf43
|
chromosome 7 open reading frame 43 |
| chr17_-_17480779 | 0.06 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr19_-_17622269 | 0.06 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr11_-_65374430 | 0.06 |
ENST00000532507.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr4_-_176733897 | 0.06 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr4_+_109571800 | 0.06 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr11_-_62995986 | 0.06 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr1_-_209957882 | 0.06 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr9_-_100000957 | 0.05 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr11_+_72975524 | 0.05 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr5_-_125930877 | 0.05 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr8_+_143781513 | 0.05 |
ENST00000292430.6
ENST00000561179.1 ENST00000518841.1 ENST00000519387.1 |
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr11_-_67211263 | 0.05 |
ENST00000393893.1
|
CORO1B
|
coronin, actin binding protein, 1B |
| chr11_+_114168085 | 0.05 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr2_-_27558270 | 0.05 |
ENST00000454704.1
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr10_+_124214164 | 0.05 |
ENST00000528446.1
|
ARMS2
|
age-related maculopathy susceptibility 2 |
| chr4_+_76439649 | 0.05 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr2_-_42160486 | 0.05 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr8_+_145215928 | 0.05 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chrX_-_107682702 | 0.05 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr2_-_208489275 | 0.05 |
ENST00000272839.3
ENST00000426075.1 |
METTL21A
|
methyltransferase like 21A |
| chr19_+_49622646 | 0.05 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr7_-_108097144 | 0.05 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_+_80267973 | 0.05 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr8_-_144660771 | 0.05 |
ENST00000449291.2
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1 |
| chr19_+_38779778 | 0.05 |
ENST00000590738.1
ENST00000587519.2 ENST00000591889.1 |
SPINT2
CTB-102L5.4
|
serine peptidase inhibitor, Kunitz type, 2 CTB-102L5.4 |
| chr12_-_48164812 | 0.05 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr3_-_4793274 | 0.05 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr10_+_134232503 | 0.04 |
ENST00000428814.1
|
RP11-432J24.2
|
RP11-432J24.2 |
| chr19_+_13134772 | 0.04 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr16_-_28621312 | 0.04 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr17_+_16284399 | 0.04 |
ENST00000535788.1
|
UBB
|
ubiquitin B |
| chr6_+_15246501 | 0.04 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr9_+_123970052 | 0.04 |
ENST00000373823.3
|
GSN
|
gelsolin |
| chr1_+_15802594 | 0.04 |
ENST00000375910.3
|
CELA2B
|
chymotrypsin-like elastase family, member 2B |
| chr6_+_134274322 | 0.04 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr3_-_9921934 | 0.04 |
ENST00000423850.1
|
CIDEC
|
cell death-inducing DFFA-like effector c |
| chr1_-_25291475 | 0.04 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr4_-_141074123 | 0.04 |
ENST00000502696.1
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr11_+_46740730 | 0.04 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr4_+_140586922 | 0.04 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr11_+_72975559 | 0.04 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr5_+_150157444 | 0.04 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr19_-_35085490 | 0.03 |
ENST00000379204.2
|
SCGB2B2
|
secretoglobin, family 2B, member 2 |
| chr7_-_139763521 | 0.03 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr12_+_118573663 | 0.03 |
ENST00000261313.2
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr1_+_15783222 | 0.03 |
ENST00000359621.4
|
CELA2A
|
chymotrypsin-like elastase family, member 2A |
| chr17_-_77005860 | 0.03 |
ENST00000591773.1
ENST00000588611.1 ENST00000586916.2 ENST00000592033.1 ENST00000588075.1 ENST00000302345.2 ENST00000591811.1 |
CANT1
|
calcium activated nucleotidase 1 |
| chr12_-_121477039 | 0.03 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr17_-_77005801 | 0.03 |
ENST00000392446.5
|
CANT1
|
calcium activated nucleotidase 1 |
| chr17_+_7253667 | 0.03 |
ENST00000570504.1
ENST00000574499.1 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr11_+_64808368 | 0.03 |
ENST00000531072.1
ENST00000398846.1 |
SAC3D1
|
SAC3 domain containing 1 |
| chr11_-_31391276 | 0.03 |
ENST00000452803.1
|
DCDC1
|
doublecortin domain containing 1 |
| chr1_-_1624083 | 0.03 |
ENST00000378662.1
ENST00000234800.6 |
SLC35E2B
|
solute carrier family 35, member E2B |
| chr11_-_111741994 | 0.03 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr16_+_69958887 | 0.03 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_+_16284104 | 0.03 |
ENST00000577958.1
ENST00000302182.3 ENST00000577640.1 |
UBB
|
ubiquitin B |
| chr17_-_41116454 | 0.03 |
ENST00000427569.2
ENST00000430739.1 |
AARSD1
|
alanyl-tRNA synthetase domain containing 1 |
| chr1_-_151032040 | 0.03 |
ENST00000540998.1
ENST00000357235.5 |
CDC42SE1
|
CDC42 small effector 1 |
| chr9_+_92219919 | 0.03 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr14_-_105071083 | 0.03 |
ENST00000415614.2
|
TMEM179
|
transmembrane protein 179 |
| chr9_-_127358087 | 0.03 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr8_+_17434689 | 0.03 |
ENST00000398074.3
|
PDGFRL
|
platelet-derived growth factor receptor-like |
| chr4_-_40516560 | 0.03 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr8_+_22844995 | 0.03 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr1_-_201140673 | 0.03 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr11_-_59578202 | 0.03 |
ENST00000300151.4
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr7_+_26331541 | 0.03 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr7_+_134233840 | 0.03 |
ENST00000457545.2
|
AKR1B15
|
aldo-keto reductase family 1, member B15 |
| chr5_+_143584814 | 0.03 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr6_-_32095968 | 0.03 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chrX_-_52533295 | 0.02 |
ENST00000375578.1
ENST00000396497.3 |
XAGE1D
|
X antigen family, member 1D |
| chr12_+_56546223 | 0.02 |
ENST00000550443.1
ENST00000207437.5 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr11_+_72975578 | 0.02 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr4_+_109571740 | 0.02 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chrX_+_52238810 | 0.02 |
ENST00000437949.2
ENST00000375616.1 |
XAGE1B
|
X antigen family, member 1B |
| chr11_+_114549108 | 0.02 |
ENST00000389586.4
ENST00000375475.5 |
NXPE2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr19_-_4457776 | 0.02 |
ENST00000301281.6
|
UBXN6
|
UBX domain protein 6 |
| chr3_+_39424828 | 0.02 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr9_+_19049372 | 0.02 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr8_-_134309335 | 0.02 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr13_-_47471155 | 0.02 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr4_-_113207048 | 0.02 |
ENST00000361717.3
|
TIFA
|
TRAF-interacting protein with forkhead-associated domain |
| chr10_+_102295616 | 0.02 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr2_+_202937972 | 0.02 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr1_-_72748417 | 0.02 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr6_-_32806483 | 0.02 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr19_-_49622348 | 0.02 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr12_-_109221160 | 0.02 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chrX_+_52511761 | 0.02 |
ENST00000399795.3
ENST00000375589.1 |
XAGE1C
|
X antigen family, member 1C |
| chr2_-_136875712 | 0.02 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chrX_-_52260355 | 0.02 |
ENST00000375602.1
ENST00000399800.3 |
XAGE1A
|
X antigen family, member 1A |
| chr20_-_44993012 | 0.02 |
ENST00000372229.1
ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr7_-_16872932 | 0.02 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr8_+_12808834 | 0.02 |
ENST00000400069.3
|
KIAA1456
|
KIAA1456 |
| chr20_+_277737 | 0.02 |
ENST00000382352.3
|
ZCCHC3
|
zinc finger, CCHC domain containing 3 |
| chr12_+_16064258 | 0.02 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr7_-_22259845 | 0.02 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chrX_+_66764375 | 0.02 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr16_-_28621298 | 0.02 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_+_122181529 | 0.02 |
ENST00000541467.1
|
TMEM120B
|
transmembrane protein 120B |
| chr2_-_208489707 | 0.02 |
ENST00000448007.2
ENST00000432416.1 ENST00000411432.1 |
METTL21A
|
methyltransferase like 21A |
| chr12_-_113574028 | 0.02 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr6_-_32806506 | 0.02 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr17_-_77005813 | 0.02 |
ENST00000590370.1
ENST00000591625.1 |
CANT1
|
calcium activated nucleotidase 1 |
| chr10_+_33271469 | 0.02 |
ENST00000414157.1
|
RP11-462L8.1
|
RP11-462L8.1 |
| chr11_+_20385231 | 0.02 |
ENST00000530266.1
ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr9_-_119162885 | 0.02 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr8_-_116680833 | 0.02 |
ENST00000220888.5
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_+_99797636 | 0.02 |
ENST00000409145.1
|
MRPL30
|
mitochondrial ribosomal protein L30 |
| chr8_-_116681123 | 0.02 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr12_-_121476959 | 0.02 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr2_-_122042770 | 0.02 |
ENST00000263707.5
|
TFCP2L1
|
transcription factor CP2-like 1 |
| chr18_-_53089723 | 0.02 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr4_+_169575875 | 0.02 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_47630255 | 0.01 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chrX_-_52546189 | 0.01 |
ENST00000375570.1
ENST00000429372.2 |
XAGE1E
|
X antigen family, member 1E |
| chr11_-_17410629 | 0.01 |
ENST00000526912.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr12_-_95044309 | 0.01 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr7_-_86595190 | 0.01 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr14_+_104394770 | 0.01 |
ENST00000409874.4
ENST00000339063.5 |
TDRD9
|
tudor domain containing 9 |
| chr1_+_21880560 | 0.01 |
ENST00000425315.2
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr8_-_116681221 | 0.01 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_-_49755019 | 0.01 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr4_-_84030996 | 0.01 |
ENST00000411416.2
|
PLAC8
|
placenta-specific 8 |
| chr17_+_66521936 | 0.01 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_-_85419603 | 0.01 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr1_+_90308981 | 0.01 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr12_+_121570631 | 0.01 |
ENST00000546057.1
ENST00000377162.2 ENST00000328963.5 ENST00000535250.1 ENST00000541446.1 |
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr1_-_153585539 | 0.01 |
ENST00000368706.4
|
S100A16
|
S100 calcium binding protein A16 |
| chr8_-_623547 | 0.01 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chrX_-_75005054 | 0.01 |
ENST00000373359.2
|
MAGEE2
|
melanoma antigen family E, 2 |
| chr1_-_155145721 | 0.01 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr2_+_102972363 | 0.01 |
ENST00000409599.1
|
IL18R1
|
interleukin 18 receptor 1 |
| chr8_+_12809093 | 0.01 |
ENST00000528753.2
|
KIAA1456
|
KIAA1456 |
| chr1_+_244816371 | 0.01 |
ENST00000263831.7
|
DESI2
|
desumoylating isopeptidase 2 |
| chr18_-_60986962 | 0.01 |
ENST00000333681.4
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr1_+_21877753 | 0.01 |
ENST00000374832.1
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr7_+_26331678 | 0.01 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr1_+_162039558 | 0.01 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr1_-_208084729 | 0.01 |
ENST00000310833.7
ENST00000356522.4 |
CD34
|
CD34 molecule |
| chr11_+_12115543 | 0.01 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr2_-_21022818 | 0.01 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr18_-_32957260 | 0.01 |
ENST00000587422.1
ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396
|
zinc finger protein 396 |
| chr12_-_16762802 | 0.01 |
ENST00000534946.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr11_+_20385327 | 0.01 |
ENST00000451739.2
ENST00000532505.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr12_-_118406028 | 0.01 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr2_+_47630108 | 0.01 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060927 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.0 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |