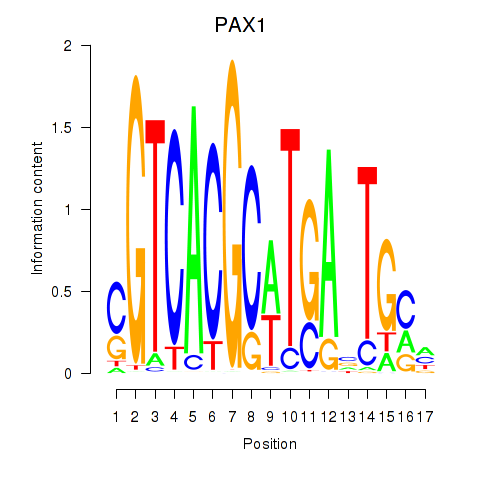
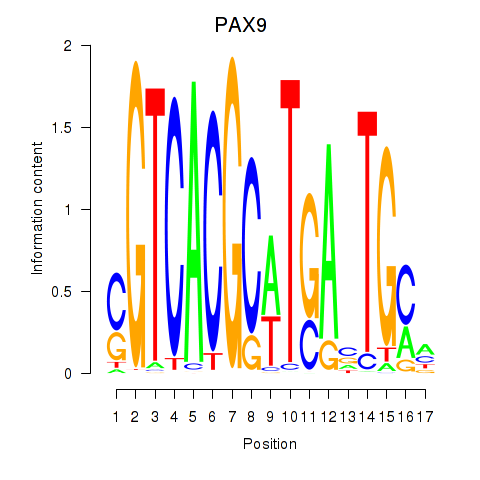
|
chrX_-_153707246
Show fit
|
0.20 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3
|
|
chrX_-_153707545
Show fit
|
0.19 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3
|
|
chr5_-_133747551
Show fit
|
0.18 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like
|
|
chr18_-_47814032
Show fit
|
0.10 |
ENST00000589548.1
ENST00000591474.1
|
CXXC1
|
CXXC finger protein 1
|
|
chr14_-_69261310
Show fit
|
0.10 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1
|
|
chr8_-_23282797
Show fit
|
0.10 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr11_-_67397371
Show fit
|
0.10 |
ENST00000376693.2
ENST00000301490.4
|
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8
|
|
chr8_-_23282820
Show fit
|
0.08 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr5_+_55354822
Show fit
|
0.08 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1
|
|
chr16_+_67197288
Show fit
|
0.07 |
ENST00000264009.8
ENST00000421453.1
|
HSF4
|
heat shock transcription factor 4
|
|
chr5_+_133842243
Show fit
|
0.06 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2
|
|
chr18_-_47813940
Show fit
|
0.06 |
ENST00000586837.1
ENST00000412036.2
ENST00000589940.1
|
CXXC1
|
CXXC finger protein 1
|
|
chr22_+_39745930
Show fit
|
0.05 |
ENST00000318801.4
ENST00000216155.7
ENST00000406293.3
ENST00000328933.5
|
SYNGR1
|
synaptogyrin 1
|
|
chr7_-_112758589
Show fit
|
0.05 |
ENST00000413744.1
ENST00000439551.1
ENST00000441359.1
|
LINC00998
|
long intergenic non-protein coding RNA 998
|
|
chr17_-_7137582
Show fit
|
0.05 |
ENST00000575756.1
ENST00000575458.1
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr5_-_133747589
Show fit
|
0.05 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like
|
|
chr9_-_131418944
Show fit
|
0.04 |
ENST00000419989.1
ENST00000451652.1
ENST00000372715.2
|
WDR34
|
WD repeat domain 34
|
|
chr6_-_170893268
Show fit
|
0.04 |
ENST00000538195.1
|
PDCD2
|
programmed cell death 2
|
|
chr12_-_122658746
Show fit
|
0.03 |
ENST00000377035.1
|
IL31
|
interleukin 31
|
|
chr7_-_112758665
Show fit
|
0.03 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998
|
|
chr17_+_39969183
Show fit
|
0.03 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr17_-_39968855
Show fit
|
0.03 |
ENST00000355468.3
ENST00000590496.1
|
LEPREL4
|
leprecan-like 4
|
|
chr15_+_63569785
Show fit
|
0.02 |
ENST00000380343.4
ENST00000560353.1
|
APH1B
|
APH1B gamma secretase subunit
|
|
chr14_+_39703112
Show fit
|
0.02 |
ENST00000555143.1
ENST00000280082.3
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr8_-_121824374
Show fit
|
0.02 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr19_-_18995029
Show fit
|
0.02 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1
|
|
chr17_-_39968406
Show fit
|
0.02 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4
|
|
chr7_-_19157248
Show fit
|
0.02 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1
|
|
chr11_+_61522844
Show fit
|
0.02 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor
|
|
chr6_+_159291090
Show fit
|
0.02 |
ENST00000367073.4
ENST00000608817.1
|
C6orf99
|
chromosome 6 open reading frame 99
|
|
chr6_+_159290917
Show fit
|
0.02 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99
|
|
chr9_-_104249400
Show fit
|
0.02 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246
|
|
chr2_-_207630033
Show fit
|
0.02 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble)
|
|
chr10_-_99393208
Show fit
|
0.02 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4
|
|
chr19_-_40791302
Show fit
|
0.02 |
ENST00000392038.2
ENST00000578123.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr19_-_39104556
Show fit
|
0.01 |
ENST00000423454.2
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr20_-_17641097
Show fit
|
0.01 |
ENST00000246043.4
|
RRBP1
|
ribosome binding protein 1
|
|
chr6_-_170893669
Show fit
|
0.01 |
ENST00000392090.2
ENST00000542896.1
ENST00000453163.2
ENST00000537445.1
|
PDCD2
|
programmed cell death 2
|
|
chr17_+_39968926
Show fit
|
0.01 |
ENST00000585664.1
ENST00000585922.1
ENST00000429461.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr18_-_47018769
Show fit
|
0.01 |
ENST00000583637.1
ENST00000578528.1
ENST00000578532.1
ENST00000580387.1
ENST00000579248.1
ENST00000581373.1
|
RPL17
|
ribosomal protein L17
|
|
chr9_+_6716478
Show fit
|
0.01 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3
|
|
chr1_+_26758790
Show fit
|
0.01 |
ENST00000427245.2
ENST00000525682.2
ENST00000236342.7
ENST00000526219.1
ENST00000374185.3
ENST00000360009.2
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr1_+_26759295
Show fit
|
0.01 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr17_+_76374714
Show fit
|
0.01 |
ENST00000262764.6
ENST00000589689.1
ENST00000329897.7
ENST00000592043.1
ENST00000587356.1
|
PGS1
|
phosphatidylglycerophosphate synthase 1
|
|
chr22_-_41940404
Show fit
|
0.01 |
ENST00000355209.4
ENST00000337566.5
ENST00000396504.2
ENST00000407461.1
|
POLR3H
|
polymerase (RNA) III (DNA directed) polypeptide H (22.9kD)
|
|
chr10_-_99393242
Show fit
|
0.01 |
ENST00000370635.3
ENST00000478953.1
ENST00000335628.3
|
MORN4
|
MORN repeat containing 4
|
|
chr19_-_40791211
Show fit
|
0.01 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr7_-_22234381
Show fit
|
0.01 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr10_-_43904608
Show fit
|
0.01 |
ENST00000337970.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chrX_+_148622513
Show fit
|
0.01 |
ENST00000393985.3
ENST00000423421.1
ENST00000423540.2
ENST00000434353.2
ENST00000514208.1
|
CXorf40A
|
chromosome X open reading frame 40A
|
|
chr17_-_3461092
Show fit
|
0.01 |
ENST00000301365.4
ENST00000572519.1
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr2_+_234160217
Show fit
|
0.01 |
ENST00000392017.4
ENST00000347464.5
ENST00000444735.1
ENST00000373525.5
ENST00000419681.1
|
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae)
|
|
chr9_+_33750667
Show fit
|
0.01 |
ENST00000457896.1
ENST00000342836.4
ENST00000429677.3
|
PRSS3
|
protease, serine, 3
|
|
chr19_-_409134
Show fit
|
0.01 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr11_+_63870660
Show fit
|
0.01 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr7_-_2349579
Show fit
|
0.01 |
ENST00000435060.1
|
SNX8
|
sorting nexin 8
|
|
chr1_+_45140360
Show fit
|
0.00 |
ENST00000418644.1
ENST00000458657.2
ENST00000441519.1
ENST00000535358.1
ENST00000445071.1
|
C1orf228
|
chromosome 1 open reading frame 228
|
|
chr3_+_127771212
Show fit
|
0.00 |
ENST00000243253.3
ENST00000481210.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr15_+_63569731
Show fit
|
0.00 |
ENST00000261879.5
|
APH1B
|
APH1B gamma secretase subunit
|
|
chr6_-_137366163
Show fit
|
0.00 |
ENST00000367748.1
|
IL20RA
|
interleukin 20 receptor, alpha
|
|
chr17_+_15848231
Show fit
|
0.00 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor
|
|
chr9_+_33795533
Show fit
|
0.00 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3
|