Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
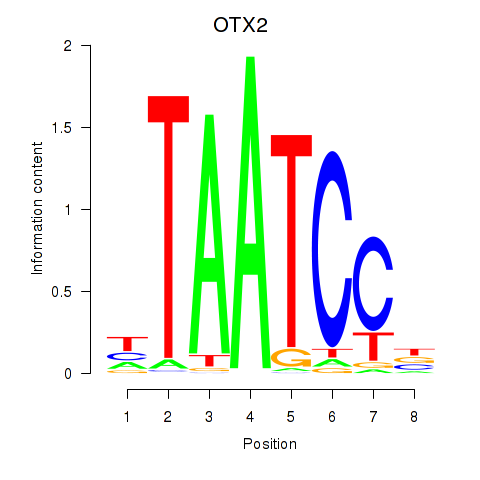
Results for OTX2_CRX
Z-value: 0.35
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CRX | hg19_v2_chr19_+_48325097_48325211 | -0.03 | 9.7e-01 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_74494779 | 0.30 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr3_-_149388682 | 0.28 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr14_-_95236551 | 0.20 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr7_-_97881429 | 0.18 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr3_+_49058444 | 0.15 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_46001697 | 0.14 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr16_-_31076332 | 0.14 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr16_-_31076273 | 0.14 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr10_+_104178946 | 0.14 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr15_-_44092295 | 0.13 |
ENST00000249714.3
ENST00000299969.6 ENST00000319327.6 |
SERINC4
|
serine incorporator 4 |
| chr1_+_200863949 | 0.13 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr18_-_56985776 | 0.11 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr3_+_111393501 | 0.11 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_+_61522844 | 0.11 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr7_-_100171270 | 0.11 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr15_+_44092784 | 0.10 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr12_+_53693466 | 0.10 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr6_+_100054606 | 0.10 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr19_+_39989535 | 0.10 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr9_-_131486367 | 0.10 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr19_+_56166360 | 0.09 |
ENST00000308924.4
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr17_-_43210580 | 0.09 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr6_-_31697563 | 0.08 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr15_-_74495188 | 0.08 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr12_-_54779511 | 0.08 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chrM_+_8366 | 0.08 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr3_-_49058479 | 0.08 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr17_-_4643161 | 0.08 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr14_-_22005343 | 0.07 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_-_31697255 | 0.07 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr10_-_126849626 | 0.07 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr3_-_54962100 | 0.07 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr11_+_61248583 | 0.07 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr19_+_39989580 | 0.07 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr3_+_49057876 | 0.07 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_50183032 | 0.06 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr2_+_219472637 | 0.06 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr7_+_28452130 | 0.06 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr17_-_43209862 | 0.06 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr19_+_7710774 | 0.06 |
ENST00000602355.1
|
STXBP2
|
syntaxin binding protein 2 |
| chr1_+_18957500 | 0.06 |
ENST00000375375.3
|
PAX7
|
paired box 7 |
| chr11_-_47521309 | 0.06 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr8_-_25281747 | 0.06 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr5_-_524445 | 0.05 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr15_+_79603479 | 0.05 |
ENST00000424155.2
ENST00000536821.1 |
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chr1_+_180897269 | 0.05 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr9_+_116298778 | 0.05 |
ENST00000462143.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chrX_-_124097620 | 0.05 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr1_-_231560790 | 0.05 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr1_+_55505184 | 0.05 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr18_-_64271363 | 0.05 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr11_+_128563652 | 0.05 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_35310312 | 0.05 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr9_-_39288092 | 0.05 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr16_-_325910 | 0.05 |
ENST00000359740.5
ENST00000316163.5 ENST00000431291.2 ENST00000397770.3 ENST00000397768.3 |
RGS11
|
regulator of G-protein signaling 11 |
| chr1_-_39395165 | 0.04 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr3_-_47950745 | 0.04 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr12_-_49523896 | 0.04 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr6_-_33297013 | 0.04 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr1_+_144989309 | 0.04 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr1_+_244214577 | 0.04 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr12_-_67197760 | 0.04 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr12_+_40787194 | 0.04 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr19_-_44124019 | 0.04 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr15_+_79603404 | 0.04 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chrX_+_15767971 | 0.04 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr1_-_150208291 | 0.04 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_161014731 | 0.04 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr17_+_74536164 | 0.04 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr3_-_137851220 | 0.04 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr14_-_22005197 | 0.04 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_+_111580508 | 0.04 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr9_-_35754253 | 0.04 |
ENST00000436428.2
|
MSMP
|
microseminoprotein, prostate associated |
| chr22_+_25615489 | 0.04 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr11_+_20044096 | 0.04 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr13_-_49975632 | 0.03 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr14_+_58894404 | 0.03 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr19_-_44123734 | 0.03 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr17_-_48207157 | 0.03 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr4_-_102268628 | 0.03 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_-_42931567 | 0.03 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr13_-_36050819 | 0.03 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr6_-_30658745 | 0.03 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr8_+_86999516 | 0.03 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr1_-_150208320 | 0.03 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_+_124735282 | 0.03 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr7_-_17500294 | 0.03 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr2_+_128175997 | 0.03 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr19_+_11457175 | 0.03 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr1_+_207277632 | 0.03 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr12_+_53693812 | 0.03 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr11_+_101983176 | 0.03 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr7_-_104909435 | 0.03 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr5_+_150040403 | 0.03 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr1_+_164528437 | 0.03 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr7_-_128171123 | 0.03 |
ENST00000608477.1
|
RP11-212P7.2
|
RP11-212P7.2 |
| chr13_-_95131923 | 0.03 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr22_+_41258250 | 0.03 |
ENST00000544094.1
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr17_+_68100989 | 0.03 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_+_50649302 | 0.03 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr3_-_50605077 | 0.03 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr1_+_11751748 | 0.03 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr4_-_153601136 | 0.03 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr11_+_72975524 | 0.03 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr9_-_95244781 | 0.03 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr10_+_24528108 | 0.03 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr11_-_8680383 | 0.03 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr21_-_19775973 | 0.03 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chrX_+_153769446 | 0.03 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr14_-_22005018 | 0.02 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr9_-_127358087 | 0.02 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr10_+_118349920 | 0.02 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr2_+_120770581 | 0.02 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr17_-_14683517 | 0.02 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr18_+_616672 | 0.02 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr1_+_153651078 | 0.02 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chrX_-_101771645 | 0.02 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chr8_-_110656995 | 0.02 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr14_+_96722152 | 0.02 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr2_-_70780572 | 0.02 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr4_+_96012585 | 0.02 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr3_-_50605150 | 0.02 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr2_-_200715573 | 0.02 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr19_-_11457162 | 0.02 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr6_+_106535455 | 0.02 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr18_+_616711 | 0.02 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr5_-_88179302 | 0.02 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr11_-_75017734 | 0.02 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr6_-_88411911 | 0.02 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr10_+_129785574 | 0.02 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr20_+_4702548 | 0.02 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr1_+_17248418 | 0.02 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr10_-_94003003 | 0.02 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_-_129592700 | 0.02 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr3_+_49059038 | 0.02 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr12_+_104697504 | 0.02 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr19_+_18208603 | 0.02 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr5_-_88178964 | 0.02 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_72975559 | 0.02 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_-_4689727 | 0.02 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chrX_+_153769409 | 0.02 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr4_+_154125565 | 0.02 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr14_-_101034407 | 0.02 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr5_+_50678921 | 0.02 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr15_-_65407524 | 0.02 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr17_+_43318434 | 0.02 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr10_-_21806759 | 0.02 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr1_+_162351503 | 0.02 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr2_+_66662510 | 0.02 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr5_-_88179017 | 0.02 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chrX_-_153363188 | 0.02 |
ENST00000303391.6
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr10_+_102505468 | 0.02 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr1_+_164528616 | 0.02 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_207277590 | 0.02 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr21_+_33245548 | 0.02 |
ENST00000270112.2
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr1_+_114522049 | 0.02 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr10_+_129785536 | 0.02 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr5_+_140345820 | 0.02 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr13_+_36050881 | 0.02 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr14_-_57197224 | 0.01 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr10_-_76868931 | 0.01 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr4_+_129730839 | 0.01 |
ENST00000511647.1
|
PHF17
|
jade family PHD finger 1 |
| chr2_-_214016314 | 0.01 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_+_129731074 | 0.01 |
ENST00000512960.1
ENST00000503785.1 ENST00000514740.1 |
PHF17
|
jade family PHD finger 1 |
| chr9_-_139361503 | 0.01 |
ENST00000453963.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr12_-_21487829 | 0.01 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr11_-_71752571 | 0.01 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_-_112218378 | 0.01 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr19_+_40877583 | 0.01 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr2_-_230787879 | 0.01 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr2_-_200320768 | 0.01 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chrX_-_100129128 | 0.01 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr11_+_72975578 | 0.01 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr2_+_173792893 | 0.01 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_62437486 | 0.01 |
ENST00000528115.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chrX_-_119077695 | 0.01 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr10_+_103986085 | 0.01 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr1_+_167298281 | 0.01 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chrM_+_10758 | 0.01 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_+_141105705 | 0.01 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_-_105845536 | 0.01 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr10_-_105845674 | 0.01 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr11_-_128894053 | 0.01 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr9_-_35072585 | 0.01 |
ENST00000448530.1
|
VCP
|
valosin containing protein |
| chr22_-_43398442 | 0.01 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr15_+_63050785 | 0.01 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr4_+_129730779 | 0.01 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr16_+_81272287 | 0.01 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr14_-_22005062 | 0.01 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr4_-_102267953 | 0.01 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chrX_-_54384425 | 0.01 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr15_+_96869165 | 0.01 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_+_133757995 | 0.01 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr2_+_33661382 | 0.01 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr15_-_52483566 | 0.01 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr17_-_39661849 | 0.01 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr7_+_136553370 | 0.01 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr7_-_111424506 | 0.01 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr20_-_31124186 | 0.01 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr10_-_49482907 | 0.01 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |