Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
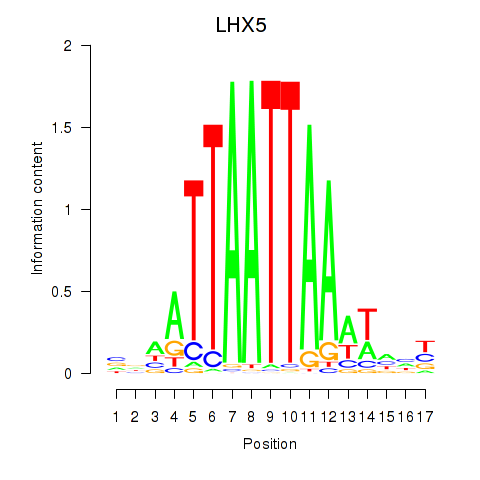
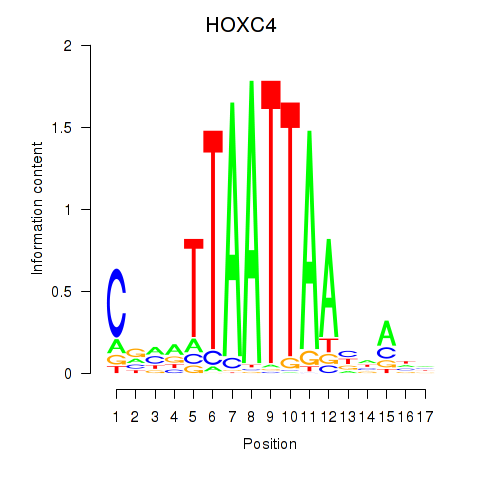
Results for OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Z-value: 1.64
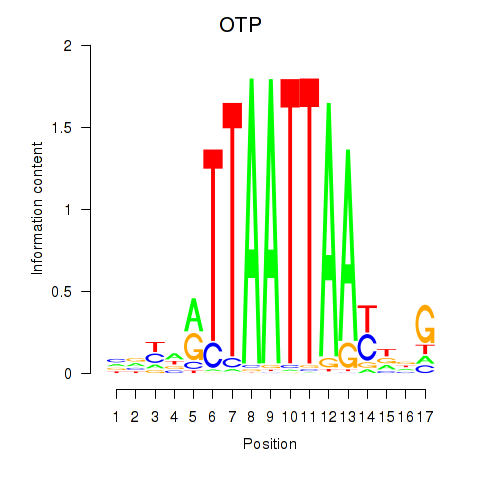
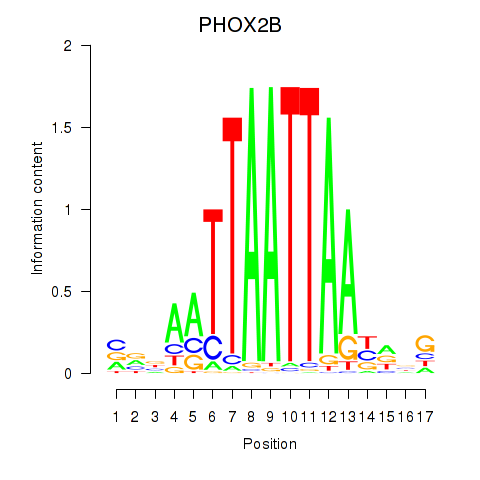
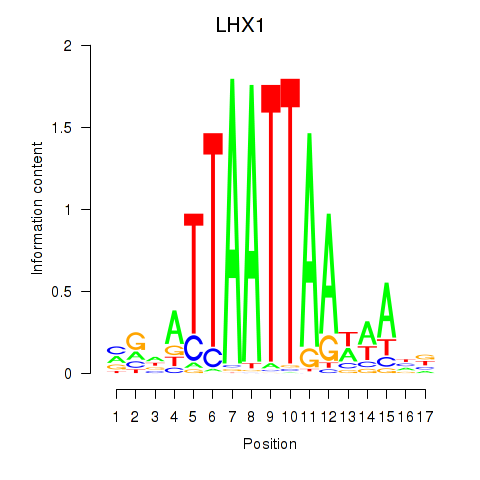
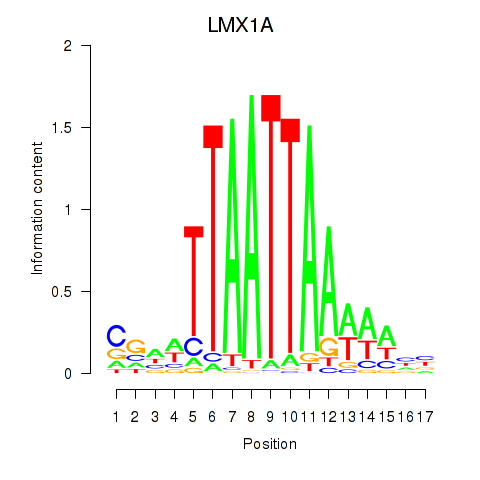
Transcription factors associated with OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTP
|
ENSG00000171540.6 | orthopedia homeobox |
|
PHOX2B
|
ENSG00000109132.5 | paired like homeobox 2B |
|
LHX1
|
ENSG00000132130.7 | LIM homeobox 1 |
|
LMX1A
|
ENSG00000162761.10 | LIM homeobox transcription factor 1 alpha |
|
LHX5
|
ENSG00000089116.3 | LIM homeobox 5 |
|
HOXC4
|
ENSG00000198353.6 | homeobox C4 |
|
HOXC4
|
ENSG00000273266.1 | homeobox C4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC4 | hg19_v2_chr12_+_54410664_54410715 | 0.85 | 1.5e-01 | Click! |
| LHX1 | hg19_v2_chr17_+_35294075_35294102 | -0.72 | 2.8e-01 | Click! |
| OTP | hg19_v2_chr5_-_76935513_76935513 | 0.33 | 6.7e-01 | Click! |
Activity profile of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Sorted Z-values of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_101417919 | 2.11 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr5_-_126409159 | 1.86 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr14_+_20187174 | 1.82 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr11_+_71934962 | 1.68 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr7_+_5465382 | 1.24 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr11_+_327171 | 0.79 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr1_+_225600404 | 0.78 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr14_+_31046959 | 0.77 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr12_-_64062583 | 0.74 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans) |
| chr4_+_183370146 | 0.65 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr11_-_327537 | 0.64 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_+_5710919 | 0.62 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr3_-_141747950 | 0.57 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_+_26402517 | 0.57 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr16_-_66583994 | 0.53 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr7_-_22862406 | 0.52 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr19_+_49199209 | 0.52 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr6_-_47445214 | 0.50 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr12_-_10978957 | 0.49 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr14_-_51027838 | 0.48 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr5_+_66300464 | 0.47 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_3259148 | 0.47 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr16_-_28937027 | 0.46 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr4_+_119810134 | 0.45 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr4_+_155484103 | 0.44 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr17_-_6524159 | 0.43 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr1_+_117544366 | 0.42 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr8_-_124741451 | 0.41 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr12_-_54653313 | 0.40 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr4_+_119809984 | 0.39 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr4_+_26324474 | 0.38 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_-_66584059 | 0.37 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr17_+_4692230 | 0.37 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr1_-_168464875 | 0.35 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr19_-_3557570 | 0.35 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr19_+_50016411 | 0.35 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chrX_+_10126488 | 0.34 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr2_-_37068530 | 0.34 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr16_+_12059091 | 0.33 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr3_+_138340049 | 0.33 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_+_38710555 | 0.32 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr16_-_66583701 | 0.32 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr12_-_46121554 | 0.31 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr17_+_74463650 | 0.31 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr4_+_155484155 | 0.30 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr7_+_138915102 | 0.30 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr17_+_57233087 | 0.30 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr4_+_113568207 | 0.30 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr6_+_114178512 | 0.29 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr10_+_24755416 | 0.28 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr5_+_40841410 | 0.28 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr14_-_75530693 | 0.28 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr1_+_151253991 | 0.27 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr19_+_50016610 | 0.26 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr20_-_50418947 | 0.26 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_+_75303993 | 0.25 |
ENST00000564779.1
|
SCAMP5
|
secretory carrier membrane protein 5 |
| chr6_-_32157947 | 0.24 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr19_-_36304201 | 0.22 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_60933634 | 0.21 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr13_-_81801115 | 0.21 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr5_+_175288631 | 0.21 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr3_+_186692745 | 0.21 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr14_+_102276192 | 0.21 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_150421752 | 0.20 |
ENST00000498386.1
|
FAM194A
|
family with sequence similarity 194, member A |
| chrX_+_43515467 | 0.19 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr4_-_177116772 | 0.19 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr10_+_6779326 | 0.19 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr4_+_169418255 | 0.19 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_10022735 | 0.19 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_112614506 | 0.18 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr12_-_118796971 | 0.18 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr7_-_144435985 | 0.17 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr5_+_179135246 | 0.17 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr17_-_38821373 | 0.17 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr4_-_74486109 | 0.17 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr16_-_71842706 | 0.17 |
ENST00000563104.1
ENST00000569975.1 ENST00000565412.1 ENST00000567583.1 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chrX_+_47696337 | 0.17 |
ENST00000334937.4
ENST00000376950.4 |
ZNF81
|
zinc finger protein 81 |
| chr17_+_68164752 | 0.16 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr2_+_152214098 | 0.16 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr7_-_22862448 | 0.16 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr15_+_58702742 | 0.16 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_+_30589829 | 0.16 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr15_-_34635314 | 0.15 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr17_+_48823975 | 0.15 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr20_-_50418972 | 0.15 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr12_+_26348582 | 0.15 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr14_-_21994337 | 0.15 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr7_+_133812052 | 0.15 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr4_-_120243545 | 0.15 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr3_+_186353756 | 0.15 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr6_+_26440700 | 0.14 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr17_+_48823896 | 0.14 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr9_-_77567743 | 0.14 |
ENST00000376854.5
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr4_+_87857538 | 0.14 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr1_+_157963063 | 0.13 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr6_-_136788001 | 0.13 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr1_-_211307404 | 0.13 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr10_-_27529486 | 0.13 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr3_+_139063372 | 0.13 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr11_-_128894053 | 0.13 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_9994021 | 0.13 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr8_-_145754428 | 0.13 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr18_-_51750948 | 0.13 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr19_+_48112371 | 0.13 |
ENST00000594866.1
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1 |
| chr12_-_118797475 | 0.13 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr14_-_80678512 | 0.12 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr5_-_53115506 | 0.12 |
ENST00000511953.1
ENST00000504552.1 |
CTD-2081C10.1
|
CTD-2081C10.1 |
| chr19_+_56280507 | 0.12 |
ENST00000341750.4
|
RFPL4AL1
|
ret finger protein-like 4A-like 1 |
| chr4_+_76481258 | 0.12 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr11_-_559377 | 0.12 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr21_-_16125773 | 0.12 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr18_+_56806701 | 0.12 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr3_+_138340067 | 0.11 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chrX_+_55744228 | 0.11 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr4_+_186990298 | 0.10 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr11_-_8857248 | 0.10 |
ENST00000534248.1
ENST00000530959.1 ENST00000531578.1 ENST00000533225.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr8_+_23145594 | 0.10 |
ENST00000519952.1
ENST00000518840.1 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
| chr9_-_131486367 | 0.10 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr4_+_26344754 | 0.10 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_-_20575959 | 0.10 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr6_+_26087646 | 0.10 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr17_-_72772462 | 0.10 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr9_+_124329336 | 0.10 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr2_-_43266680 | 0.10 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chrX_+_84258832 | 0.10 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr14_-_39639523 | 0.10 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr2_+_234826016 | 0.10 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr1_-_233431458 | 0.10 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr16_-_28634874 | 0.10 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_+_56270380 | 0.10 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr5_+_95066823 | 0.09 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr18_-_47018869 | 0.09 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr3_-_149293990 | 0.09 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_+_67498538 | 0.09 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr7_+_134528635 | 0.09 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr1_+_66820058 | 0.09 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr4_+_71019903 | 0.09 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr5_+_140762268 | 0.09 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr17_+_66511540 | 0.09 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr6_+_26087509 | 0.09 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr10_+_91152303 | 0.09 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr21_+_33671160 | 0.09 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr6_-_138833630 | 0.08 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr3_-_33686925 | 0.08 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_-_74619152 | 0.08 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chrM_-_14670 | 0.08 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr15_-_55489097 | 0.08 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr13_+_46276441 | 0.08 |
ENST00000310521.1
ENST00000533564.1 |
SPERT
|
spermatid associated |
| chr4_+_169418195 | 0.08 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_-_77225135 | 0.08 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr13_+_24144796 | 0.07 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_13677795 | 0.07 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr1_-_153518270 | 0.07 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr1_-_39347255 | 0.07 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr10_-_4285923 | 0.07 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr7_-_143599207 | 0.07 |
ENST00000355951.2
ENST00000479870.1 ENST00000478172.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr12_+_4130143 | 0.07 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr16_-_71842941 | 0.07 |
ENST00000423132.2
ENST00000433195.2 ENST00000569748.1 ENST00000570017.1 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr4_+_75174180 | 0.07 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr15_+_93443419 | 0.07 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr7_+_13141097 | 0.07 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr12_-_95510743 | 0.07 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr19_+_36139125 | 0.07 |
ENST00000246554.3
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr6_+_26402465 | 0.07 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr3_-_185538849 | 0.06 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_-_98417780 | 0.06 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr3_-_141719195 | 0.06 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_12510352 | 0.06 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr2_+_102953608 | 0.06 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr1_-_190446759 | 0.06 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_-_24645956 | 0.06 |
ENST00000543707.1
|
KIAA0319
|
KIAA0319 |
| chr9_-_215744 | 0.06 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr2_+_102456277 | 0.06 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr8_-_95487272 | 0.06 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr6_-_41039567 | 0.06 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr3_+_39424828 | 0.06 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr1_+_12806141 | 0.06 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr9_-_99540328 | 0.06 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr12_+_26348429 | 0.05 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr14_+_50234309 | 0.05 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr20_-_50419055 | 0.05 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr10_-_46620012 | 0.05 |
ENST00000508602.1
ENST00000374339.3 ENST00000502254.1 ENST00000437863.1 ENST00000374342.2 ENST00000395722.3 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr3_+_20081515 | 0.05 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr5_+_40841276 | 0.05 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr12_-_11287243 | 0.05 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr8_-_29605625 | 0.05 |
ENST00000506121.3
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr20_+_15177480 | 0.05 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr1_-_109935819 | 0.05 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr14_+_72052983 | 0.05 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_+_54466179 | 0.04 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr2_+_102615416 | 0.04 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr6_-_133035185 | 0.04 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr14_-_104387888 | 0.04 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr1_-_101360205 | 0.04 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr9_-_26947220 | 0.04 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr7_+_13141010 | 0.04 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr16_-_71843047 | 0.04 |
ENST00000299980.4
ENST00000393512.3 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr4_-_36245561 | 0.04 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_95640218 | 0.04 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr2_+_166095898 | 0.04 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 1.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.2 | 0.6 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.7 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.7 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.5 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 1.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.0 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.2 | 2.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.7 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |