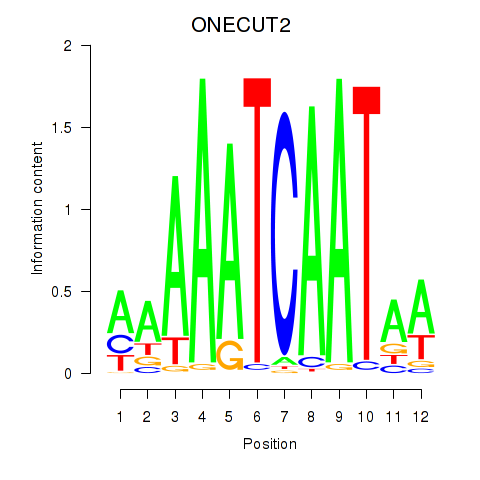
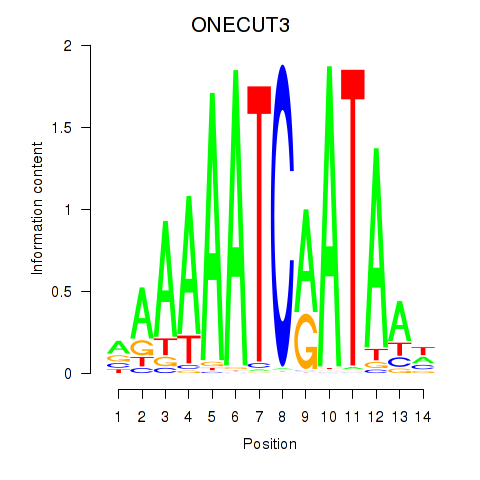
Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ONECUT2_ONECUT3
Z-value: 0.54
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | -0.97 | 3.5e-02 | Click! |
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | -0.28 | 7.2e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_142264664 | 0.62 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr2_-_192016276 | 0.40 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr2_-_208030295 | 0.31 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_147052 | 0.31 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr12_-_58212487 | 0.27 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr11_+_8040739 | 0.26 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr3_+_187461442 | 0.26 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr17_-_46692457 | 0.22 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr15_-_83837983 | 0.21 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr6_+_126221034 | 0.17 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_-_4535233 | 0.17 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr8_+_133787586 | 0.16 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr12_-_91546926 | 0.14 |
ENST00000550758.1
|
DCN
|
decorin |
| chr7_-_100239132 | 0.14 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr6_+_56954808 | 0.14 |
ENST00000510483.1
ENST00000370706.4 ENST00000357489.3 |
ZNF451
|
zinc finger protein 451 |
| chr1_+_207943667 | 0.14 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr3_+_159557637 | 0.13 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_+_64846129 | 0.13 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chrX_-_119693745 | 0.13 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr2_-_113012592 | 0.13 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
| chr13_-_36429763 | 0.12 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr12_+_64845864 | 0.12 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr2_-_192016316 | 0.12 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr7_-_28220354 | 0.12 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr7_-_151330218 | 0.12 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr5_-_119669160 | 0.12 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr1_-_198906528 | 0.11 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr1_-_26233423 | 0.11 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr17_+_61151306 | 0.11 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr16_-_75467274 | 0.10 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr2_+_102413726 | 0.10 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_-_93747425 | 0.10 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr8_+_120885949 | 0.10 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr7_-_30009542 | 0.10 |
ENST00000438497.1
|
SCRN1
|
secernin 1 |
| chr12_-_54653313 | 0.10 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr11_-_47736896 | 0.09 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr16_-_66952779 | 0.09 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr14_+_93357749 | 0.09 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr13_+_31774073 | 0.09 |
ENST00000343307.4
|
B3GALTL
|
beta 1,3-galactosyltransferase-like |
| chr4_-_103747011 | 0.08 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_71632894 | 0.08 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr16_-_28192360 | 0.08 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr16_-_66952742 | 0.08 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr9_+_70856397 | 0.08 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chrX_-_85302531 | 0.08 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr18_+_11752040 | 0.08 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_+_171561127 | 0.08 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr6_-_151773232 | 0.07 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr9_-_70490107 | 0.07 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr4_-_103746924 | 0.07 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_+_86098670 | 0.07 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr17_-_37764128 | 0.07 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr4_-_102268708 | 0.07 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr11_+_28724129 | 0.07 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr5_+_136070614 | 0.07 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr10_+_114710516 | 0.07 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_55646412 | 0.07 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_-_116383738 | 0.07 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr12_+_59989918 | 0.07 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr15_+_93443419 | 0.07 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr2_-_39348137 | 0.07 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr10_-_46167722 | 0.07 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr1_-_100598444 | 0.07 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr5_-_143550241 | 0.06 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr10_+_133753533 | 0.06 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr3_-_11685345 | 0.06 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr2_+_109204743 | 0.06 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_103746683 | 0.06 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_27925019 | 0.06 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr16_+_72459838 | 0.06 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr3_-_141747439 | 0.06 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_+_62164340 | 0.06 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr1_+_223354486 | 0.06 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr4_+_71588372 | 0.06 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_+_34643600 | 0.06 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr12_+_86268065 | 0.06 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr1_-_53686261 | 0.06 |
ENST00000294360.4
|
C1orf123
|
chromosome 1 open reading frame 123 |
| chr17_+_44701402 | 0.06 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr5_+_162887556 | 0.06 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr7_-_26240357 | 0.05 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr4_+_71587669 | 0.05 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_+_43380459 | 0.05 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr2_+_67624430 | 0.05 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr11_-_116658695 | 0.05 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr17_+_79369249 | 0.05 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr12_-_122018114 | 0.05 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr17_-_39623681 | 0.05 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr12_+_32832203 | 0.05 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chrX_+_70586082 | 0.05 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr6_+_56954919 | 0.05 |
ENST00000508603.1
ENST00000491832.2 ENST00000370710.6 |
ZNF451
|
zinc finger protein 451 |
| chr1_+_207262578 | 0.05 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr12_+_32832134 | 0.05 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr19_-_44952635 | 0.05 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr6_+_71122974 | 0.05 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr17_-_46691990 | 0.05 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr2_-_208030886 | 0.05 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_48866448 | 0.05 |
ENST00000266594.1
|
ANP32D
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member D |
| chr11_-_118550375 | 0.04 |
ENST00000525958.1
ENST00000264029.4 ENST00000397925.1 ENST00000529101.1 |
TREH
|
trehalase (brush-border membrane glycoprotein) |
| chr11_-_116658758 | 0.04 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr11_+_6947647 | 0.04 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr3_-_100565249 | 0.04 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_-_186080012 | 0.04 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr10_-_121296045 | 0.04 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr3_+_130650738 | 0.04 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr12_+_2912363 | 0.04 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr14_+_77607026 | 0.04 |
ENST00000600936.1
|
AC007375.1
|
Uncharacterized protein; cDNA FLJ43210 fis, clone FEBRA2020582 |
| chr3_-_141747459 | 0.04 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_+_142829162 | 0.04 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr11_-_107729887 | 0.04 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_-_75467318 | 0.04 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr4_+_71248795 | 0.04 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chrX_+_100805496 | 0.04 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chr2_-_86850949 | 0.04 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr8_-_117043 | 0.04 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr1_+_145883868 | 0.04 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr10_-_73975657 | 0.03 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr8_+_22601 | 0.03 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr1_-_115259337 | 0.03 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr5_-_146833222 | 0.03 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr5_+_65440032 | 0.03 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chrX_+_123095890 | 0.03 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr22_-_36220420 | 0.03 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_100925967 | 0.03 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr15_+_38746307 | 0.03 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr17_-_76824975 | 0.03 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr14_+_19377522 | 0.03 |
ENST00000550708.1
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12 |
| chr9_-_27529726 | 0.03 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr5_-_156390230 | 0.03 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr11_-_10828892 | 0.03 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr4_+_156824840 | 0.03 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr5_+_138940742 | 0.03 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr1_-_29557383 | 0.03 |
ENST00000373791.3
ENST00000263702.6 |
MECR
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr1_-_147610081 | 0.03 |
ENST00000369226.3
|
NBPF24
|
neuroblastoma breakpoint family, member 24 |
| chr4_+_174818390 | 0.03 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr12_-_4488872 | 0.03 |
ENST00000237837.1
|
FGF23
|
fibroblast growth factor 23 |
| chr2_+_109204909 | 0.03 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_28443799 | 0.03 |
ENST00000584423.1
ENST00000247026.5 |
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr7_-_23510086 | 0.02 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr2_+_58655461 | 0.02 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr7_+_99425633 | 0.02 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr11_-_102651343 | 0.02 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr12_-_10601963 | 0.02 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chrX_+_123095860 | 0.02 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr5_+_147774275 | 0.02 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr10_+_69869237 | 0.02 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chrX_-_64196351 | 0.02 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_+_76352178 | 0.02 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr1_+_99127225 | 0.02 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr20_-_1974692 | 0.02 |
ENST00000217305.2
ENST00000539905.1 |
PDYN
|
prodynorphin |
| chr9_+_71944241 | 0.02 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_-_76817036 | 0.02 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr17_+_28705921 | 0.02 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chrX_-_64196376 | 0.02 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr2_+_170440902 | 0.02 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr5_-_143550159 | 0.02 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr7_+_43152212 | 0.02 |
ENST00000453890.1
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr12_+_9144626 | 0.02 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr7_+_116451100 | 0.02 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr12_+_100594557 | 0.02 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr1_+_110655050 | 0.02 |
ENST00000334179.3
|
UBL4B
|
ubiquitin-like 4B |
| chrX_+_46771848 | 0.02 |
ENST00000218343.4
|
PHF16
|
jade family PHD finger 3 |
| chr16_-_69788816 | 0.02 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr2_-_220174166 | 0.01 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chrX_-_19988382 | 0.01 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr4_-_184241927 | 0.01 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr22_+_46449674 | 0.01 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr20_+_36405665 | 0.01 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr1_+_207262540 | 0.01 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr2_+_157330081 | 0.01 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr14_+_39736582 | 0.01 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chrX_+_27608490 | 0.01 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr7_+_133615169 | 0.01 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr10_-_419129 | 0.01 |
ENST00000540204.1
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr16_-_28374829 | 0.01 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chrX_-_117119243 | 0.01 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr3_+_69985734 | 0.01 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_-_111150048 | 0.01 |
ENST00000485317.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chrX_+_46771711 | 0.01 |
ENST00000424392.1
ENST00000397189.1 |
PHF16
|
jade family PHD finger 3 |
| chr11_+_85359062 | 0.01 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr14_-_24047965 | 0.01 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr11_-_60623437 | 0.01 |
ENST00000332539.4
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr1_+_207262627 | 0.01 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr7_+_129007964 | 0.01 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_-_146068184 | 0.01 |
ENST00000604894.1
ENST00000369323.3 ENST00000479926.2 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr11_+_134144139 | 0.01 |
ENST00000389887.5
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr2_-_32490859 | 0.01 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr1_-_243326612 | 0.01 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr14_+_67831576 | 0.01 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr9_+_77230499 | 0.01 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr7_-_78400598 | 0.01 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_37131058 | 0.01 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr6_-_79787902 | 0.01 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr10_-_62332357 | 0.01 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr7_-_14026063 | 0.00 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr2_+_190541153 | 0.00 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr5_+_128300810 | 0.00 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr4_+_169418195 | 0.00 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr15_-_37392724 | 0.00 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr2_-_50201327 | 0.00 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr17_-_67264947 | 0.00 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |