Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
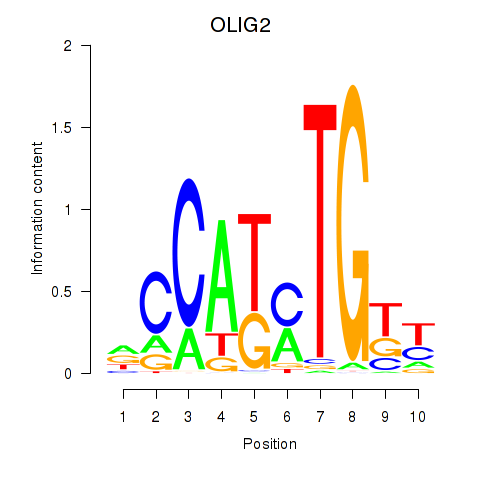
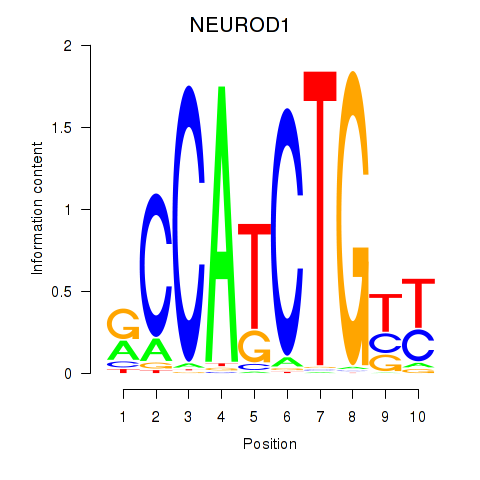
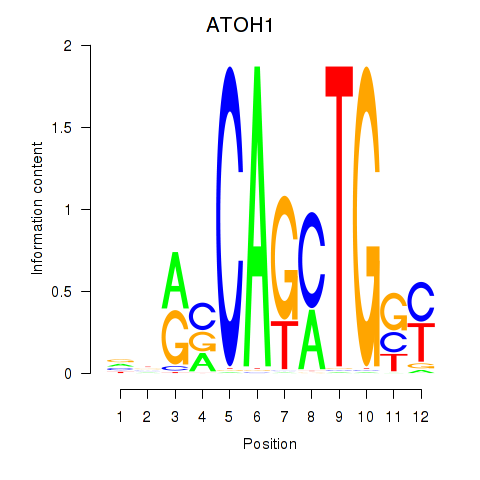
Results for OLIG2_NEUROD1_ATOH1
Z-value: 0.81
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.4 | oligodendrocyte transcription factor 2 |
|
NEUROD1
|
ENSG00000162992.3 | neuronal differentiation 1 |
|
ATOH1
|
ENSG00000172238.3 | atonal bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROD1 | hg19_v2_chr2_-_182545603_182545603 | -0.85 | 1.5e-01 | Click! |
| ATOH1 | hg19_v2_chr4_+_94750014_94750042 | -0.59 | 4.1e-01 | Click! |
| OLIG2 | hg19_v2_chr21_+_34398153_34398250 | -0.03 | 9.7e-01 | Click! |
Activity profile of OLIG2_NEUROD1_ATOH1 motif
Sorted Z-values of OLIG2_NEUROD1_ATOH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_149785236 | 0.65 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr15_-_94614049 | 0.64 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr22_-_21905120 | 0.53 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr14_+_104710541 | 0.43 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr6_-_33663474 | 0.43 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr3_+_72201910 | 0.38 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr7_-_150777920 | 0.36 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr1_-_149812765 | 0.36 |
ENST00000369158.1
|
HIST2H3C
|
histone cluster 2, H3c |
| chr3_-_99833333 | 0.35 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_-_123752624 | 0.35 |
ENST00000542174.1
ENST00000535796.1 |
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr8_+_39972170 | 0.34 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr1_-_226076843 | 0.32 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr22_+_45072925 | 0.32 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr12_-_52715179 | 0.30 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr6_-_26250835 | 0.30 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr20_-_31124186 | 0.30 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr17_+_39421591 | 0.29 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr18_-_56985776 | 0.28 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr19_-_19030157 | 0.26 |
ENST00000349893.4
ENST00000351079.4 ENST00000600932.1 ENST00000262812.4 |
COPE
|
coatomer protein complex, subunit epsilon |
| chr14_+_21359558 | 0.25 |
ENST00000304639.3
|
RNASE3
|
ribonuclease, RNase A family, 3 |
| chrX_+_47444613 | 0.25 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr22_-_42342692 | 0.25 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr5_+_139055021 | 0.24 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr9_+_131174024 | 0.24 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr3_-_178789220 | 0.24 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr1_-_171621815 | 0.23 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr3_-_52868931 | 0.23 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr9_+_19408919 | 0.22 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr9_+_135037334 | 0.22 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr7_-_150777949 | 0.22 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr8_-_135522425 | 0.22 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr11_+_827553 | 0.22 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr17_+_33474826 | 0.21 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr22_+_45072958 | 0.21 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr11_+_76494253 | 0.21 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr10_+_18429606 | 0.21 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr7_-_150777874 | 0.21 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_+_75931861 | 0.20 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr1_-_74663825 | 0.20 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr11_-_14913765 | 0.19 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr7_+_143078379 | 0.19 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr10_-_8095412 | 0.19 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr16_+_77246337 | 0.19 |
ENST00000563157.1
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_-_11450249 | 0.19 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr17_+_42925270 | 0.18 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr8_-_22014255 | 0.18 |
ENST00000424267.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr19_-_39368887 | 0.18 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr10_-_13043697 | 0.18 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr15_+_75660431 | 0.18 |
ENST00000563278.1
|
RP11-817O13.8
|
RP11-817O13.8 |
| chr16_+_2510081 | 0.18 |
ENST00000361837.4
ENST00000569496.1 ENST00000567489.1 ENST00000563531.1 ENST00000483320.1 |
C16orf59
|
chromosome 16 open reading frame 59 |
| chr17_-_27405875 | 0.17 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr1_-_63782888 | 0.17 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr13_+_102104980 | 0.17 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_+_122150646 | 0.17 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr15_-_75660919 | 0.16 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr18_+_59415396 | 0.16 |
ENST00000567801.1
|
RP11-1096D5.1
|
RP11-1096D5.1 |
| chr2_-_220117867 | 0.16 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr6_+_159290917 | 0.16 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr21_+_30502806 | 0.16 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr14_+_97925151 | 0.16 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr11_-_57417405 | 0.16 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr19_-_49944806 | 0.16 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr1_-_39339777 | 0.16 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr10_+_102759545 | 0.15 |
ENST00000454422.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr17_+_74723031 | 0.15 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr3_-_64253655 | 0.15 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr5_+_139927213 | 0.15 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr4_+_42399856 | 0.15 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr12_+_48152774 | 0.15 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr17_-_41132088 | 0.15 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr8_-_22014339 | 0.15 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr19_-_14992264 | 0.14 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr16_+_30662050 | 0.14 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr1_-_33647267 | 0.14 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr1_+_155051305 | 0.14 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr12_-_49318715 | 0.14 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr18_-_56985873 | 0.14 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr19_+_41222998 | 0.14 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr18_+_18943554 | 0.14 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr1_+_180165672 | 0.14 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr17_+_76164639 | 0.14 |
ENST00000225777.3
ENST00000585591.1 ENST00000589711.1 ENST00000588282.1 ENST00000589168.1 |
SYNGR2
|
synaptogyrin 2 |
| chr8_-_123706338 | 0.14 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr9_+_2017063 | 0.14 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_50183032 | 0.14 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr14_+_101359265 | 0.14 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr16_+_30662085 | 0.14 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr13_-_88323514 | 0.14 |
ENST00000441617.1
|
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr19_-_55791058 | 0.14 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr7_-_74867509 | 0.14 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr3_-_11623804 | 0.13 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_-_226111929 | 0.13 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr8_+_67405794 | 0.13 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chrX_+_135279179 | 0.13 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_77681075 | 0.13 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chr17_+_76165213 | 0.13 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr5_-_176326333 | 0.13 |
ENST00000292432.5
|
HK3
|
hexokinase 3 (white cell) |
| chrX_+_100474763 | 0.13 |
ENST00000395209.3
|
DRP2
|
dystrophin related protein 2 |
| chr18_+_43914159 | 0.13 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr10_-_15413035 | 0.13 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr14_-_75078725 | 0.13 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr19_-_4454081 | 0.13 |
ENST00000591919.1
|
UBXN6
|
UBX domain protein 6 |
| chr19_-_46285646 | 0.13 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr15_+_75940218 | 0.12 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr4_+_53525573 | 0.12 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr7_+_143078652 | 0.12 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chrX_+_70521584 | 0.12 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr4_-_24914576 | 0.12 |
ENST00000502801.1
ENST00000428116.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr4_+_103422499 | 0.12 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr11_-_71639613 | 0.12 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr16_+_57847684 | 0.12 |
ENST00000335616.2
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr2_+_196440692 | 0.11 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_-_18480260 | 0.11 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr19_-_56056888 | 0.11 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr10_-_64576105 | 0.11 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr12_+_57916466 | 0.11 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr15_+_89182156 | 0.11 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr3_-_87040259 | 0.11 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr12_-_130529501 | 0.11 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr14_+_38065052 | 0.11 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr5_+_139055055 | 0.11 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr19_+_54695098 | 0.11 |
ENST00000396388.2
|
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr12_-_120805872 | 0.11 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chrX_+_135278908 | 0.11 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr2_+_11052054 | 0.11 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr1_+_74663896 | 0.11 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr1_+_159141397 | 0.11 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr11_+_46299199 | 0.11 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr12_+_116997186 | 0.11 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr19_+_36249044 | 0.10 |
ENST00000444637.2
ENST00000396908.4 ENST00000544099.1 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr12_+_53846594 | 0.10 |
ENST00000550192.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr19_-_1650666 | 0.10 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr17_+_7477040 | 0.10 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr22_+_42372970 | 0.10 |
ENST00000291236.11
|
SEPT3
|
septin 3 |
| chr11_+_66115304 | 0.10 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr14_+_32414059 | 0.10 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr1_+_28261533 | 0.10 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_+_117073850 | 0.09 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr4_+_90033968 | 0.09 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr13_-_26625169 | 0.09 |
ENST00000319420.3
|
SHISA2
|
shisa family member 2 |
| chr11_-_73694346 | 0.09 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr14_-_21562648 | 0.09 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr3_+_49058444 | 0.09 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr17_-_42452063 | 0.09 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr7_+_72848092 | 0.09 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr12_+_58176525 | 0.09 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr19_-_6501778 | 0.09 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr11_-_66675371 | 0.09 |
ENST00000393955.2
|
PC
|
pyruvate carboxylase |
| chr21_+_30503282 | 0.09 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_-_798239 | 0.09 |
ENST00000531437.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr2_-_166930131 | 0.09 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr16_+_21716284 | 0.09 |
ENST00000388957.3
|
OTOA
|
otoancorin |
| chr17_-_19651598 | 0.09 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr13_-_20767037 | 0.09 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr6_+_30851840 | 0.09 |
ENST00000511510.1
ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_47546220 | 0.09 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr2_-_7005785 | 0.08 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr11_+_45918092 | 0.08 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_-_49058479 | 0.08 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr1_-_149889382 | 0.08 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr3_-_45267760 | 0.08 |
ENST00000503771.1
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene) |
| chr4_-_54424366 | 0.08 |
ENST00000306888.2
|
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr17_+_14277419 | 0.08 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chr15_-_63448973 | 0.08 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr17_+_33474860 | 0.08 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr12_-_57328187 | 0.08 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_+_30174426 | 0.08 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_101039402 | 0.08 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr20_+_55205825 | 0.08 |
ENST00000544508.1
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr7_+_30174574 | 0.08 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr2_-_42160486 | 0.08 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chrX_+_118108571 | 0.08 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr3_-_172859017 | 0.08 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chr2_-_239148599 | 0.08 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr11_-_71639670 | 0.08 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr7_-_97881429 | 0.08 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr6_-_41909466 | 0.08 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr17_-_73892992 | 0.08 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr12_+_53491220 | 0.08 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr1_-_161993422 | 0.08 |
ENST00000367940.2
|
OLFML2B
|
olfactomedin-like 2B |
| chr15_-_78526855 | 0.07 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr5_-_133747551 | 0.07 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr17_-_79166176 | 0.07 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr17_-_46623441 | 0.07 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr17_+_65040678 | 0.07 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr12_-_58146048 | 0.07 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr19_-_46285736 | 0.07 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chr11_-_2170786 | 0.07 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr10_+_102758105 | 0.07 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr7_+_143079000 | 0.07 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr2_+_95963052 | 0.07 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr20_-_39946237 | 0.07 |
ENST00000441102.2
ENST00000559234.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chrX_-_151619746 | 0.07 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr18_-_53068782 | 0.07 |
ENST00000569012.1
|
TCF4
|
transcription factor 4 |
| chr2_+_85661918 | 0.07 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr1_+_10271674 | 0.07 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr2_+_69001913 | 0.07 |
ENST00000409030.3
ENST00000409220.1 |
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_+_16062820 | 0.07 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr17_+_73089382 | 0.07 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr16_+_71660079 | 0.07 |
ENST00000565261.1
ENST00000268485.3 ENST00000299952.4 |
MARVELD3
|
MARVEL domain containing 3 |
| chr19_+_19030478 | 0.07 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG2_NEUROD1_ATOH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.2 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.0 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.0 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |