Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
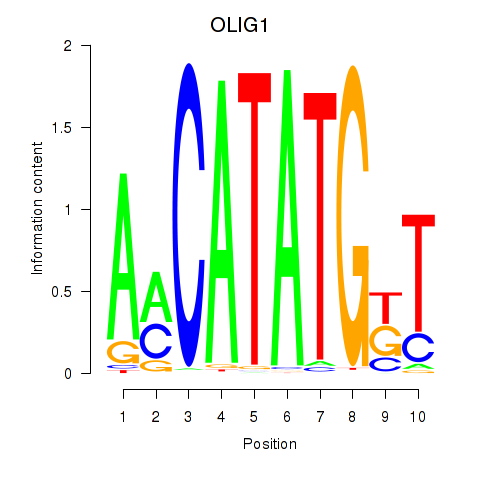
Results for OLIG1
Z-value: 0.43
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG1
|
ENSG00000184221.8 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OLIG1 | hg19_v2_chr21_+_34442439_34442455 | 0.58 | 4.2e-01 | Click! |
Activity profile of OLIG1 motif
Sorted Z-values of OLIG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_75168071 | 0.46 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr3_-_129375556 | 0.37 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr6_+_24126350 | 0.33 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr11_-_44972418 | 0.30 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr17_-_73874654 | 0.23 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr1_-_44818599 | 0.21 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr7_-_112758589 | 0.21 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr7_+_4721885 | 0.20 |
ENST00000328914.4
|
FOXK1
|
forkhead box K1 |
| chr9_-_35079911 | 0.20 |
ENST00000448890.1
|
FANCG
|
Fanconi anemia, complementation group G |
| chr11_-_62359027 | 0.19 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr1_+_81106951 | 0.18 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr7_+_140396465 | 0.18 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr1_-_186344802 | 0.18 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr17_+_60758814 | 0.17 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr16_+_68279207 | 0.17 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr11_-_44972476 | 0.17 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr18_-_46784778 | 0.17 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr16_-_14726576 | 0.17 |
ENST00000562896.1
|
PARN
|
poly(A)-specific ribonuclease |
| chr16_+_2933209 | 0.16 |
ENST00000293981.6
|
FLYWCH2
|
FLYWCH family member 2 |
| chr2_-_166930131 | 0.16 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr11_-_62358972 | 0.16 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr14_-_23822080 | 0.15 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr19_-_39368887 | 0.15 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr4_-_159094194 | 0.15 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr1_-_16563641 | 0.15 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr19_-_54974894 | 0.15 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr16_+_68279256 | 0.15 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr7_+_140396756 | 0.15 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr1_-_8000872 | 0.15 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr18_+_44812072 | 0.14 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr16_+_2933229 | 0.14 |
ENST00000573965.1
ENST00000572006.1 |
FLYWCH2
|
FLYWCH family member 2 |
| chr15_+_40650408 | 0.14 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr16_-_30905584 | 0.14 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr3_+_72201910 | 0.14 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr2_+_220462560 | 0.14 |
ENST00000456909.1
ENST00000295641.10 |
STK11IP
|
serine/threonine kinase 11 interacting protein |
| chr18_-_47814032 | 0.14 |
ENST00000589548.1
ENST00000591474.1 |
CXXC1
|
CXXC finger protein 1 |
| chr22_+_20877924 | 0.14 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr19_+_42041860 | 0.13 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr20_-_32031680 | 0.13 |
ENST00000217381.2
|
SNTA1
|
syntrophin, alpha 1 |
| chr11_-_26743546 | 0.13 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr15_-_41166414 | 0.13 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr16_+_12059091 | 0.13 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr12_+_7052974 | 0.12 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr17_-_17485731 | 0.12 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr7_-_112758665 | 0.12 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr19_+_40873617 | 0.12 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr19_-_11450249 | 0.12 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr19_+_18496957 | 0.12 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr2_-_37501692 | 0.11 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr3_-_49466686 | 0.11 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr7_-_44530479 | 0.11 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr15_+_75940218 | 0.11 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr10_+_5238793 | 0.11 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr3_-_148939598 | 0.10 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr22_+_38054721 | 0.10 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr7_+_127527965 | 0.10 |
ENST00000486037.1
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr17_-_7145475 | 0.10 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr18_+_3411595 | 0.10 |
ENST00000552383.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_203274639 | 0.10 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr2_+_96068436 | 0.10 |
ENST00000445649.1
ENST00000447036.1 ENST00000233379.4 ENST00000418606.1 |
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr12_-_123450986 | 0.10 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr16_-_69385681 | 0.10 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr1_-_231560790 | 0.09 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr6_+_163148161 | 0.09 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr18_-_47813940 | 0.09 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr19_-_1021113 | 0.09 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr14_+_23344345 | 0.09 |
ENST00000551466.1
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr16_+_2933187 | 0.09 |
ENST00000396958.3
|
FLYWCH2
|
FLYWCH family member 2 |
| chr17_+_49243639 | 0.09 |
ENST00000512737.1
ENST00000503064.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr4_+_144354644 | 0.09 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr20_+_34556488 | 0.09 |
ENST00000373973.3
ENST00000349339.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr19_+_46498704 | 0.08 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr16_-_57798253 | 0.08 |
ENST00000565270.1
|
KIFC3
|
kinesin family member C3 |
| chr11_-_6633799 | 0.08 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr8_+_39972170 | 0.08 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr6_+_4087664 | 0.08 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chrX_-_84363974 | 0.08 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr13_+_28519343 | 0.08 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr12_+_117013656 | 0.08 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr2_-_97760576 | 0.08 |
ENST00000414820.1
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr9_-_132404374 | 0.08 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr22_+_47070490 | 0.08 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr7_-_48068699 | 0.08 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr7_-_72972319 | 0.08 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr1_+_244214577 | 0.08 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr16_+_31119615 | 0.08 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr2_-_183387064 | 0.07 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr22_-_19435755 | 0.07 |
ENST00000542103.1
ENST00000399562.4 |
C22orf39
|
chromosome 22 open reading frame 39 |
| chr2_-_183106641 | 0.07 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_80797886 | 0.07 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr8_-_95449155 | 0.07 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr1_-_24306835 | 0.07 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr18_-_29340827 | 0.07 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chr15_+_85144217 | 0.07 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr10_+_5135981 | 0.07 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chrX_+_120181457 | 0.07 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr1_+_149239529 | 0.07 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr6_-_28891709 | 0.07 |
ENST00000377194.3
ENST00000377199.3 |
TRIM27
|
tripartite motif containing 27 |
| chr17_+_46048497 | 0.07 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr4_+_57276661 | 0.07 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr17_+_46048471 | 0.07 |
ENST00000578018.1
ENST00000579175.1 |
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr20_+_48429233 | 0.07 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr3_-_197300194 | 0.07 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr3_-_48936272 | 0.07 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr3_+_9839335 | 0.07 |
ENST00000453882.1
|
ARPC4-TTLL3
|
ARPC4-TTLL3 readthrough |
| chr19_+_44764031 | 0.07 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr8_+_35649365 | 0.07 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr10_-_5446786 | 0.07 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr19_-_39390350 | 0.07 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr19_-_42806919 | 0.07 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr5_-_159766528 | 0.06 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr19_+_19569270 | 0.06 |
ENST00000358713.3
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr3_+_137717571 | 0.06 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr7_-_37024665 | 0.06 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr6_+_37400974 | 0.06 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr2_+_108994466 | 0.06 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr1_-_160549235 | 0.06 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr1_+_196743912 | 0.06 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr22_+_24999114 | 0.06 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr6_+_17110726 | 0.06 |
ENST00000354384.5
|
STMND1
|
stathmin domain containing 1 |
| chr1_+_12524965 | 0.06 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_15479054 | 0.06 |
ENST00000376014.3
ENST00000451326.2 |
TMEM51
|
transmembrane protein 51 |
| chr12_+_133656995 | 0.06 |
ENST00000356456.5
|
ZNF140
|
zinc finger protein 140 |
| chr8_-_135522425 | 0.06 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr1_+_76251912 | 0.05 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr1_-_205091115 | 0.05 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr2_+_11674213 | 0.05 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_+_37440653 | 0.05 |
ENST00000328376.5
ENST00000452017.2 |
C3orf35
|
chromosome 3 open reading frame 35 |
| chr1_+_206808868 | 0.05 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr1_+_17559776 | 0.05 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chr4_+_109571740 | 0.05 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr11_+_126262027 | 0.05 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr3_-_58200398 | 0.05 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr17_-_39538550 | 0.05 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr18_-_59274139 | 0.05 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr6_+_57182400 | 0.05 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr1_-_53387386 | 0.05 |
ENST00000467988.1
ENST00000358358.5 ENST00000371522.4 |
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr12_-_11175219 | 0.05 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr10_+_81462983 | 0.05 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr1_-_45253377 | 0.05 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr10_-_1071796 | 0.05 |
ENST00000277517.1
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr3_-_43147549 | 0.05 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr3_+_49840685 | 0.05 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr16_+_24550857 | 0.05 |
ENST00000568015.1
|
RBBP6
|
retinoblastoma binding protein 6 |
| chr21_+_42792442 | 0.05 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr7_+_1727755 | 0.05 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr11_-_72493574 | 0.04 |
ENST00000536290.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr3_-_128690173 | 0.04 |
ENST00000508239.1
|
RP11-723O4.6
|
Uncharacterized protein FLJ43738 |
| chr11_+_134123389 | 0.04 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chr17_+_42923686 | 0.04 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr19_+_42041702 | 0.04 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr22_+_37678505 | 0.04 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chr19_+_18208603 | 0.04 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr14_-_60632162 | 0.04 |
ENST00000557185.1
|
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr4_+_118955500 | 0.04 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr6_-_34393825 | 0.04 |
ENST00000605528.1
ENST00000326199.8 |
RPS10-NUDT3
RPS10
|
RPS10-NUDT3 readthrough ribosomal protein S10 |
| chr11_+_32154200 | 0.04 |
ENST00000525133.1
|
RP1-65P5.5
|
RP1-65P5.5 |
| chr17_+_7211280 | 0.04 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr2_+_54342574 | 0.04 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr14_+_38091270 | 0.04 |
ENST00000553443.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chrX_-_13752675 | 0.04 |
ENST00000380579.1
ENST00000458511.2 ENST00000519885.1 ENST00000358231.5 ENST00000518847.1 ENST00000453655.2 ENST00000359680.5 |
TRAPPC2
|
trafficking protein particle complex 2 |
| chr17_+_80858418 | 0.04 |
ENST00000574422.1
|
TBCD
|
tubulin folding cofactor D |
| chr5_+_140227357 | 0.04 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr1_+_180897269 | 0.04 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr8_+_101349823 | 0.04 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr18_-_44561988 | 0.04 |
ENST00000332567.4
|
TCEB3B
|
transcription elongation factor B polypeptide 3B (elongin A2) |
| chr2_+_201994208 | 0.04 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_166428839 | 0.04 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_-_115127611 | 0.03 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chrX_-_69509738 | 0.03 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr2_-_70418032 | 0.03 |
ENST00000425268.1
ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42
|
chromosome 2 open reading frame 42 |
| chr1_-_249111272 | 0.03 |
ENST00000411742.2
|
SH3BP5L
|
SH3-binding domain protein 5-like |
| chr1_+_110881945 | 0.03 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr8_-_16050142 | 0.03 |
ENST00000536385.1
ENST00000381998.4 |
MSR1
|
macrophage scavenger receptor 1 |
| chr19_+_21324827 | 0.03 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr3_-_18480260 | 0.03 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_196743943 | 0.03 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr4_-_120243545 | 0.03 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr8_+_21915368 | 0.03 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr9_-_95244781 | 0.03 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr7_-_2854860 | 0.03 |
ENST00000447791.1
ENST00000407904.3 |
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr7_-_107968921 | 0.03 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr12_-_16761007 | 0.03 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_196857144 | 0.03 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr12_+_4699244 | 0.03 |
ENST00000540757.2
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr2_-_228582709 | 0.03 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr1_+_220863187 | 0.03 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr2_-_183387430 | 0.03 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_53387352 | 0.03 |
ENST00000541281.1
|
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr16_-_420338 | 0.03 |
ENST00000450882.1
ENST00000441883.1 ENST00000447696.1 ENST00000389675.2 |
MRPL28
|
mitochondrial ribosomal protein L28 |
| chrX_+_107288197 | 0.03 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr19_+_39390320 | 0.03 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr17_+_7211656 | 0.03 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr16_-_55867146 | 0.03 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr22_-_30953587 | 0.03 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_-_98515395 | 0.03 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr14_-_60632011 | 0.03 |
ENST00000554101.1
ENST00000557137.1 |
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr5_-_115872124 | 0.03 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr22_-_42526802 | 0.03 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr11_-_111741994 | 0.03 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.0 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.5 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.0 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0098519 | phosphoserine phosphatase activity(GO:0004647) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |