Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
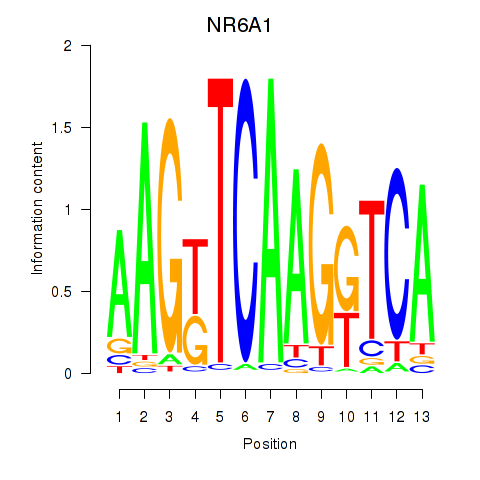
Results for NR6A1
Z-value: 0.40
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | nuclear receptor subfamily 6 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR6A1 | hg19_v2_chr9_-_127533519_127533576 | -0.58 | 4.2e-01 | Click! |
Activity profile of NR6A1 motif
Sorted Z-values of NR6A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_94536514 | 0.15 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr17_-_46799872 | 0.14 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr1_+_40997233 | 0.13 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr19_+_11670245 | 0.13 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr3_+_172034218 | 0.12 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr4_-_185776854 | 0.12 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr1_-_161207986 | 0.12 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr8_+_67104323 | 0.12 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr8_+_32579271 | 0.12 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr10_-_14574705 | 0.12 |
ENST00000489100.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr6_-_31138439 | 0.11 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr4_-_10459009 | 0.11 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr3_+_182971335 | 0.10 |
ENST00000464191.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr11_-_46141338 | 0.10 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr10_-_14613968 | 0.10 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr8_-_150563 | 0.09 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr17_-_36413133 | 0.09 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr8_+_32579321 | 0.09 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr10_-_14614095 | 0.09 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr11_-_72852320 | 0.09 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr12_+_123942188 | 0.09 |
ENST00000526639.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr10_-_14614311 | 0.08 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr10_+_114710516 | 0.08 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_10366223 | 0.08 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr1_+_197886461 | 0.08 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr10_-_14614122 | 0.08 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr1_-_1763721 | 0.08 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr10_-_75634219 | 0.08 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr7_-_130353553 | 0.08 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr16_-_59788718 | 0.08 |
ENST00000564533.1
|
RP11-105C20.2
|
Uncharacterized protein |
| chr14_-_58893832 | 0.07 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr9_+_114393581 | 0.07 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr17_+_44679808 | 0.07 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr11_+_30344595 | 0.07 |
ENST00000282032.3
|
ARL14EP
|
ADP-ribosylation factor-like 14 effector protein |
| chr8_-_97247759 | 0.07 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr19_-_43690642 | 0.07 |
ENST00000407356.1
ENST00000407568.1 ENST00000404580.1 ENST00000599812.1 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chr2_+_207024306 | 0.07 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr3_+_57882024 | 0.07 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr3_+_37284668 | 0.07 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr19_+_42381173 | 0.07 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr7_-_95225768 | 0.07 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr3_-_148804275 | 0.07 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr2_-_69180083 | 0.07 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr18_+_8609402 | 0.07 |
ENST00000329286.6
|
RAB12
|
RAB12, member RAS oncogene family |
| chr10_+_46222648 | 0.07 |
ENST00000336378.4
ENST00000540872.1 ENST00000537517.1 ENST00000374362.2 ENST00000359860.4 ENST00000420848.1 |
FAM21C
|
family with sequence similarity 21, member C |
| chr16_+_53412368 | 0.07 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr13_+_98086445 | 0.07 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr19_+_42381337 | 0.07 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr4_+_39184024 | 0.07 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr6_+_31638156 | 0.06 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr3_-_139108463 | 0.06 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr14_+_89029336 | 0.06 |
ENST00000556945.1
ENST00000556158.1 ENST00000557607.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr7_-_6048702 | 0.06 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chrX_-_48823165 | 0.06 |
ENST00000419374.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr8_-_102275131 | 0.06 |
ENST00000523121.1
|
KB-1410C5.2
|
KB-1410C5.2 |
| chr9_+_114393634 | 0.06 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr10_+_16478942 | 0.06 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr1_-_159825137 | 0.06 |
ENST00000368102.1
|
C1orf204
|
chromosome 1 open reading frame 204 |
| chr16_-_30997533 | 0.06 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr12_-_96793142 | 0.06 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr11_+_113185292 | 0.06 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr3_-_113464906 | 0.06 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr4_-_10458982 | 0.06 |
ENST00000326756.3
|
ZNF518B
|
zinc finger protein 518B |
| chr3_-_139108475 | 0.06 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr12_+_26164645 | 0.06 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr14_+_89029253 | 0.06 |
ENST00000251038.5
ENST00000359301.3 ENST00000302216.8 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr16_+_69166770 | 0.06 |
ENST00000567235.2
ENST00000568448.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr19_-_16008880 | 0.06 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr5_+_52285144 | 0.06 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr1_-_245026388 | 0.06 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr1_+_214776516 | 0.06 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr17_+_66509019 | 0.06 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr11_-_73720122 | 0.06 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr1_+_159770292 | 0.06 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr12_+_7917812 | 0.06 |
ENST00000382119.1
|
NANOGNB
|
NANOG neighbor homeobox |
| chr2_-_178128149 | 0.06 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr10_+_81107271 | 0.06 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr4_-_170948361 | 0.06 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr10_+_105726862 | 0.06 |
ENST00000335753.4
ENST00000369755.3 |
SLK
|
STE20-like kinase |
| chr6_+_31637944 | 0.06 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr15_-_32747103 | 0.05 |
ENST00000562377.1
|
GOLGA8O
|
golgin A8 family, member O |
| chr2_+_69240511 | 0.05 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr14_+_74058410 | 0.05 |
ENST00000326303.4
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr3_-_139396560 | 0.05 |
ENST00000514703.1
ENST00000511444.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr10_+_90346519 | 0.05 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr15_+_101420028 | 0.05 |
ENST00000557963.1
ENST00000346623.6 |
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr16_-_31228680 | 0.05 |
ENST00000302964.3
|
PYDC1
|
PYD (pyrin domain) containing 1 |
| chr9_+_131217459 | 0.05 |
ENST00000497812.2
ENST00000393533.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr12_+_57810198 | 0.05 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr10_+_116697946 | 0.05 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr11_+_110001723 | 0.05 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr18_+_11751493 | 0.05 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr4_-_7044657 | 0.05 |
ENST00000310085.4
|
CCDC96
|
coiled-coil domain containing 96 |
| chr20_+_43803517 | 0.05 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr11_-_111957451 | 0.05 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr1_+_212738676 | 0.05 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr19_+_53761545 | 0.05 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr14_+_89029366 | 0.05 |
ENST00000555799.1
ENST00000555755.1 ENST00000393514.5 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr9_-_21305312 | 0.05 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr11_-_16430399 | 0.05 |
ENST00000528252.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chrX_+_23682379 | 0.05 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr10_-_104866395 | 0.05 |
ENST00000458345.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr3_+_37427760 | 0.05 |
ENST00000425932.1
|
C3orf35
|
chromosome 3 open reading frame 35 |
| chr4_+_128702969 | 0.05 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr2_+_171640291 | 0.05 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr12_+_53399942 | 0.05 |
ENST00000262056.9
|
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr20_+_57209828 | 0.05 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chrX_-_118827113 | 0.05 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr11_+_113185571 | 0.05 |
ENST00000455306.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr2_+_26467825 | 0.04 |
ENST00000545822.1
ENST00000425035.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr16_-_12897642 | 0.04 |
ENST00000433677.2
ENST00000261660.4 ENST00000381774.4 |
CPPED1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr9_-_114362093 | 0.04 |
ENST00000538962.1
ENST00000407693.2 ENST00000238248.3 |
PTGR1
|
prostaglandin reductase 1 |
| chr19_-_10446449 | 0.04 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr3_-_72496035 | 0.04 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr19_+_45542773 | 0.04 |
ENST00000544944.2
|
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr17_-_79817091 | 0.04 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr13_-_114898016 | 0.04 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chrX_-_46187069 | 0.04 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr3_-_139396853 | 0.04 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr1_+_29213584 | 0.04 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr16_-_47177874 | 0.04 |
ENST00000562435.1
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr10_+_70480963 | 0.04 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr2_-_211090162 | 0.04 |
ENST00000233710.3
|
ACADL
|
acyl-CoA dehydrogenase, long chain |
| chr19_-_43244610 | 0.04 |
ENST00000595140.1
ENST00000327495.5 |
PSG3
|
pregnancy specific beta-1-glycoprotein 3 |
| chr6_+_129204337 | 0.04 |
ENST00000421865.2
|
LAMA2
|
laminin, alpha 2 |
| chr10_-_126107482 | 0.04 |
ENST00000368845.5
ENST00000539214.1 |
OAT
|
ornithine aminotransferase |
| chr5_-_86708670 | 0.04 |
ENST00000504878.1
|
CCNH
|
cyclin H |
| chr5_+_140027355 | 0.04 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr10_-_61513201 | 0.04 |
ENST00000414264.1
ENST00000594536.1 |
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr12_+_54384370 | 0.04 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chrX_-_118827333 | 0.04 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr7_+_156742399 | 0.04 |
ENST00000275820.3
|
NOM1
|
nucleolar protein with MIF4G domain 1 |
| chr10_-_61513146 | 0.04 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr1_+_111415757 | 0.04 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr1_-_161207875 | 0.04 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_161102367 | 0.04 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr7_+_142919130 | 0.04 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr17_-_37308824 | 0.04 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr9_+_131447342 | 0.04 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr5_-_14871866 | 0.04 |
ENST00000284268.6
|
ANKH
|
ANKH inorganic pyrophosphate transport regulator |
| chr5_+_34656529 | 0.04 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr7_-_75401513 | 0.04 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr14_-_75389975 | 0.04 |
ENST00000555647.1
ENST00000557413.1 |
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr20_+_33462745 | 0.04 |
ENST00000488172.1
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr13_+_23755054 | 0.04 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr2_-_231084820 | 0.04 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr5_+_34656569 | 0.04 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr19_+_49999631 | 0.04 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr1_+_156338993 | 0.04 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr2_-_26467557 | 0.04 |
ENST00000380649.3
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr12_+_121837905 | 0.04 |
ENST00000392465.3
ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34
|
ring finger protein 34, E3 ubiquitin protein ligase |
| chr2_+_219283815 | 0.04 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr2_+_54558004 | 0.04 |
ENST00000405749.1
ENST00000398634.2 ENST00000447328.1 |
C2orf73
|
chromosome 2 open reading frame 73 |
| chr10_+_85933494 | 0.04 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr19_-_43773589 | 0.04 |
ENST00000291752.5
ENST00000244293.7 ENST00000596730.1 ENST00000593948.1 ENST00000270077.3 ENST00000443718.3 ENST00000418820.2 |
PSG9
|
pregnancy specific beta-1-glycoprotein 9 |
| chr3_+_73045936 | 0.04 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr22_-_22900001 | 0.04 |
ENST00000403441.1
|
PRAME
|
preferentially expressed antigen in melanoma |
| chr6_-_31620095 | 0.03 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr2_-_3521518 | 0.03 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr15_-_54051831 | 0.03 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr3_+_185046676 | 0.03 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_33701707 | 0.03 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr4_+_113152978 | 0.03 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_101437487 | 0.03 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr4_+_113152881 | 0.03 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_69240302 | 0.03 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr19_-_43530600 | 0.03 |
ENST00000598133.1
ENST00000403486.1 ENST00000306322.7 ENST00000401740.1 |
PSG11
|
pregnancy specific beta-1-glycoprotein 11 |
| chr10_+_47894023 | 0.03 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr9_-_14314518 | 0.03 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr1_+_46379254 | 0.03 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr5_-_156486120 | 0.03 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr7_-_6048650 | 0.03 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr2_+_152266604 | 0.03 |
ENST00000430328.2
|
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr6_+_62284008 | 0.03 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr17_+_57784826 | 0.03 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr20_-_61051026 | 0.03 |
ENST00000252997.2
|
GATA5
|
GATA binding protein 5 |
| chr5_+_34757309 | 0.03 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr4_-_78740769 | 0.03 |
ENST00000512485.1
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr11_+_113185251 | 0.03 |
ENST00000529221.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr2_+_26467762 | 0.03 |
ENST00000317799.5
ENST00000405867.3 ENST00000537713.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr7_-_100844193 | 0.03 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr1_-_29508499 | 0.03 |
ENST00000373795.4
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr11_-_64013663 | 0.03 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_-_43269809 | 0.03 |
ENST00000406636.3
ENST00000404209.4 ENST00000306511.4 |
PSG8
|
pregnancy specific beta-1-glycoprotein 8 |
| chr15_-_65477637 | 0.03 |
ENST00000300107.3
|
CLPX
|
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr2_+_69240415 | 0.03 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr2_-_65593784 | 0.03 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_+_99933730 | 0.03 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr6_+_107349392 | 0.03 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr7_+_94536898 | 0.03 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr7_-_69062391 | 0.03 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr11_+_34643600 | 0.03 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr20_-_35402123 | 0.03 |
ENST00000373740.3
ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1
|
DSN1, MIS12 kinetochore complex component |
| chr9_+_15553055 | 0.03 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr22_+_40390930 | 0.03 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr10_-_70092671 | 0.03 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr14_-_55738788 | 0.03 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chrX_-_69479654 | 0.03 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr11_-_47788847 | 0.03 |
ENST00000263773.5
|
FNBP4
|
formin binding protein 4 |
| chr12_-_15865844 | 0.03 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
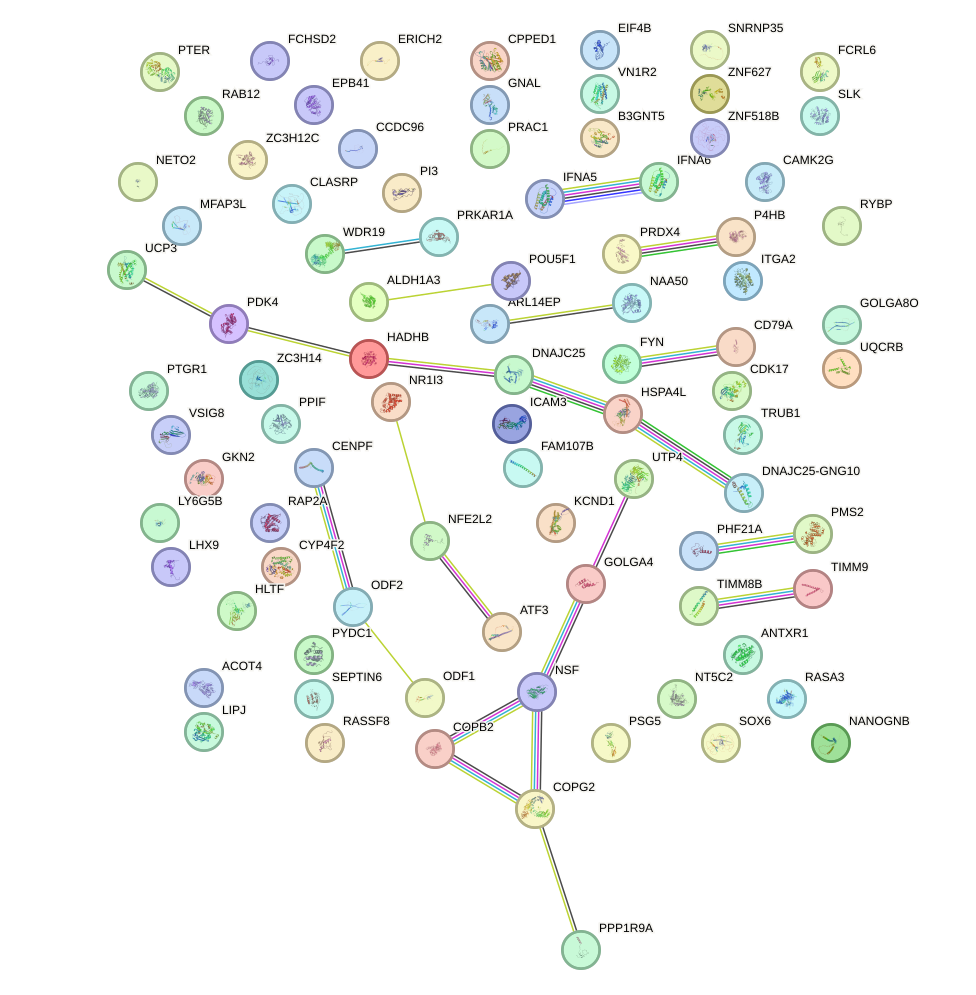
Network of associatons between targets according to the STRING database.
First level regulatory network of NR6A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.0 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.0 | GO:0019254 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.0 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |