Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
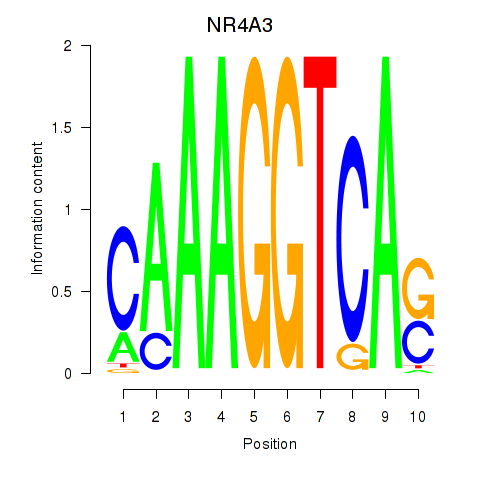
Results for NR4A3
Z-value: 0.99
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | nuclear receptor subfamily 4 group A member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | -0.64 | 3.6e-01 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_145752390 | 0.92 |
ENST00000529415.2
ENST00000533758.1 |
LRRC24
|
leucine rich repeat containing 24 |
| chr14_-_23791484 | 0.87 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr16_+_691792 | 0.81 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr19_+_6372444 | 0.60 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr6_-_43197189 | 0.52 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr11_+_66624527 | 0.52 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr8_-_131028782 | 0.51 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr11_-_45939565 | 0.51 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr15_-_75199213 | 0.51 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr11_-_45939374 | 0.50 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr11_+_64073699 | 0.49 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr1_-_12679171 | 0.47 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr7_+_7222157 | 0.46 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr17_-_7307358 | 0.45 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr19_+_34287174 | 0.42 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr8_-_145550337 | 0.42 |
ENST00000531896.1
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr7_-_105926058 | 0.40 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr4_-_1723040 | 0.38 |
ENST00000382936.3
ENST00000536901.1 ENST00000303277.2 |
TMEM129
|
transmembrane protein 129 |
| chr12_+_121163538 | 0.37 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr7_-_150777874 | 0.37 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_-_49339080 | 0.36 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr14_+_35451880 | 0.36 |
ENST00000554803.1
ENST00000555746.1 |
SRP54
|
signal recognition particle 54kDa |
| chr1_+_16085263 | 0.36 |
ENST00000483633.2
ENST00000502739.1 ENST00000431771.2 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr7_+_13141010 | 0.35 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr9_+_139746792 | 0.34 |
ENST00000317446.2
ENST00000445819.1 |
MAMDC4
|
MAM domain containing 4 |
| chr14_-_102976135 | 0.34 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr14_-_23288930 | 0.34 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr8_-_144897138 | 0.34 |
ENST00000377533.3
|
SCRIB
|
scribbled planar cell polarity protein |
| chr11_+_66059339 | 0.33 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr11_+_63742050 | 0.33 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr17_+_79679369 | 0.33 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr1_+_16083098 | 0.32 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr12_+_121163602 | 0.32 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr19_-_10444188 | 0.31 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr4_-_40632757 | 0.30 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr14_+_74035763 | 0.29 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr22_+_30821732 | 0.29 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr22_-_27620603 | 0.29 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr11_+_63993738 | 0.29 |
ENST00000441250.2
ENST00000279206.3 |
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr11_-_118272610 | 0.29 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr11_+_57508825 | 0.29 |
ENST00000534355.1
|
C11orf31
|
chromosome 11 open reading frame 31 |
| chr10_-_76995675 | 0.28 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr20_-_36156125 | 0.28 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr19_+_35810164 | 0.28 |
ENST00000598537.1
|
CD22
|
CD22 molecule |
| chr11_+_57559005 | 0.27 |
ENST00000534647.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr3_-_9994021 | 0.27 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr19_-_49552363 | 0.26 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr16_-_2581409 | 0.26 |
ENST00000567119.1
ENST00000565480.1 ENST00000382350.1 |
CEMP1
|
cementum protein 1 |
| chr7_-_74489609 | 0.26 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr17_+_73606766 | 0.26 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr11_-_61659006 | 0.26 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr4_-_159094194 | 0.26 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr17_+_79679299 | 0.26 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr3_-_50605077 | 0.25 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr7_-_150777949 | 0.25 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr22_+_50354104 | 0.25 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr2_+_121493717 | 0.25 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr3_-_145940214 | 0.25 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr1_+_156698743 | 0.24 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr15_+_59908633 | 0.24 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr11_-_117698765 | 0.24 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr20_-_36156264 | 0.24 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr16_+_2034183 | 0.24 |
ENST00000569451.1
ENST00000248114.6 ENST00000561710.1 |
GFER
|
growth factor, augmenter of liver regeneration |
| chr11_+_57509020 | 0.24 |
ENST00000388857.4
ENST00000528798.1 |
C11orf31
|
chromosome 11 open reading frame 31 |
| chr22_+_37959647 | 0.24 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr10_-_76995769 | 0.24 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr19_-_633576 | 0.24 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr11_+_67798363 | 0.23 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr16_-_67450325 | 0.23 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr17_+_36858694 | 0.23 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr16_+_28875126 | 0.22 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr7_-_17500294 | 0.22 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr22_+_30115986 | 0.22 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr15_-_74494779 | 0.22 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr2_+_128175997 | 0.22 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr6_-_31633624 | 0.22 |
ENST00000375895.2
ENST00000375900.4 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr5_-_141703713 | 0.22 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr19_+_1248547 | 0.22 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr12_+_113376249 | 0.22 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr16_+_29823552 | 0.22 |
ENST00000300797.6
|
PRRT2
|
proline-rich transmembrane protein 2 |
| chr15_+_75335604 | 0.21 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr1_+_210502238 | 0.21 |
ENST00000545154.1
ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT
|
hedgehog acyltransferase |
| chr1_-_46769261 | 0.21 |
ENST00000343304.6
|
LRRC41
|
leucine rich repeat containing 41 |
| chr12_-_6715808 | 0.21 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr15_+_29211570 | 0.21 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr22_+_29702996 | 0.21 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_-_151138323 | 0.21 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr11_-_63993690 | 0.21 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr4_-_40632881 | 0.21 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr14_-_102976091 | 0.21 |
ENST00000286918.4
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr19_+_19639670 | 0.21 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr12_+_133067157 | 0.21 |
ENST00000261673.6
|
FBRSL1
|
fibrosin-like 1 |
| chr17_+_79373540 | 0.20 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr9_-_136344237 | 0.20 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr16_+_29823427 | 0.20 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr1_+_24117627 | 0.20 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr8_-_145018905 | 0.20 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr17_+_55173955 | 0.20 |
ENST00000576591.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr2_+_98262497 | 0.20 |
ENST00000258424.2
|
COX5B
|
cytochrome c oxidase subunit Vb |
| chr16_+_2880254 | 0.20 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr19_+_49713991 | 0.20 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr22_+_45072925 | 0.19 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr2_+_120189422 | 0.19 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr19_-_15236470 | 0.19 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr7_-_642261 | 0.19 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr17_-_77924627 | 0.19 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_+_160175166 | 0.19 |
ENST00000368077.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr12_-_46121554 | 0.19 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr19_-_41256207 | 0.19 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr16_-_28937027 | 0.19 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr22_+_45072958 | 0.18 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr17_+_77893135 | 0.18 |
ENST00000574526.1
ENST00000572353.1 |
RP11-353N14.4
|
RP11-353N14.4 |
| chr11_+_35639735 | 0.18 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr2_-_206950996 | 0.18 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr19_-_15236562 | 0.18 |
ENST00000263383.3
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr1_+_113217309 | 0.18 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_-_50311896 | 0.18 |
ENST00000529634.2
|
FUZ
|
fuzzy planar cell polarity protein |
| chr19_-_3062465 | 0.18 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr9_-_140095186 | 0.18 |
ENST00000409012.4
|
TPRN
|
taperin |
| chr7_-_150777920 | 0.18 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr20_+_19738792 | 0.17 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr16_-_2168079 | 0.17 |
ENST00000488185.2
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr1_+_92495528 | 0.17 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr6_-_41909466 | 0.17 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr6_-_32143828 | 0.17 |
ENST00000412465.2
ENST00000375107.3 |
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr11_+_66824346 | 0.17 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr22_-_30960876 | 0.17 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_-_4852332 | 0.17 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr20_-_36156293 | 0.17 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr1_+_16083123 | 0.17 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr19_-_48867171 | 0.17 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr17_-_46682321 | 0.17 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr18_+_72168325 | 0.16 |
ENST00000582666.1
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr9_-_4299874 | 0.16 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chrX_+_47078069 | 0.16 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr19_-_10679644 | 0.16 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr21_+_45773515 | 0.16 |
ENST00000397932.2
ENST00000300481.9 |
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr2_-_73460334 | 0.16 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr1_+_156698708 | 0.16 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr14_-_107283278 | 0.16 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr16_+_81272287 | 0.16 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr4_-_140527848 | 0.16 |
ENST00000608795.1
ENST00000608958.1 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr11_+_66824276 | 0.16 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr11_-_117699413 | 0.16 |
ENST00000528014.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr4_-_40632140 | 0.16 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr9_-_130487143 | 0.16 |
ENST00000419060.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr19_+_10196981 | 0.15 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_+_95831529 | 0.15 |
ENST00000295210.6
ENST00000453539.2 |
ZNF2
|
zinc finger protein 2 |
| chr8_-_97247759 | 0.15 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr19_+_40697514 | 0.15 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr9_+_35749203 | 0.15 |
ENST00000456972.2
ENST00000378078.4 |
RGP1
|
RGP1 retrograde golgi transport homolog (S. cerevisiae) |
| chr18_+_3411595 | 0.15 |
ENST00000552383.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_+_140019004 | 0.15 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr20_+_1246908 | 0.15 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr12_-_109219937 | 0.15 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr16_-_73082274 | 0.15 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr6_-_41909561 | 0.15 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr12_+_53817633 | 0.15 |
ENST00000257863.4
ENST00000550311.1 ENST00000379791.3 |
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr1_-_9811600 | 0.15 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr12_+_50794947 | 0.15 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chrX_-_77225135 | 0.15 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr15_-_43622736 | 0.15 |
ENST00000544735.1
ENST00000567039.1 ENST00000305641.5 |
LCMT2
|
leucine carboxyl methyltransferase 2 |
| chr15_-_78112553 | 0.15 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr11_-_1783633 | 0.15 |
ENST00000367196.3
|
CTSD
|
cathepsin D |
| chr9_-_139839064 | 0.14 |
ENST00000325285.3
ENST00000428398.1 |
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr6_-_32144838 | 0.14 |
ENST00000395499.1
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr7_+_148892557 | 0.14 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr19_+_7011509 | 0.14 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr19_+_17416457 | 0.14 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr14_-_81408063 | 0.14 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr19_-_5624057 | 0.14 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr11_-_70507867 | 0.14 |
ENST00000412252.1
ENST00000409161.1 ENST00000409530.1 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_+_24117662 | 0.14 |
ENST00000420982.1
ENST00000374505.2 |
LYPLA2
|
lysophospholipase II |
| chr3_+_120315160 | 0.14 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr3_-_197300194 | 0.14 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr12_-_2966193 | 0.13 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr1_+_45274154 | 0.13 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr16_-_67969888 | 0.13 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr13_-_95364389 | 0.13 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr20_-_17641097 | 0.13 |
ENST00000246043.4
|
RRBP1
|
ribosome binding protein 1 |
| chr6_+_19837592 | 0.13 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr17_-_7991021 | 0.13 |
ENST00000319144.4
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chr3_-_47934234 | 0.13 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr5_+_32710736 | 0.13 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_-_226065330 | 0.13 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr19_-_51289436 | 0.13 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chrX_-_150067272 | 0.13 |
ENST00000355149.3
ENST00000437787.2 |
CD99L2
|
CD99 molecule-like 2 |
| chrX_-_150067173 | 0.12 |
ENST00000370377.3
ENST00000320893.6 |
CD99L2
|
CD99 molecule-like 2 |
| chr14_+_97059070 | 0.12 |
ENST00000553378.1
ENST00000555496.1 |
RP11-433J8.1
|
RP11-433J8.1 |
| chr22_+_50925213 | 0.12 |
ENST00000395733.3
ENST00000216075.6 ENST00000395732.3 |
MIOX
|
myo-inositol oxygenase |
| chr1_-_155232221 | 0.12 |
ENST00000355379.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chrX_+_120181457 | 0.12 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr11_-_57282349 | 0.12 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr19_-_42927251 | 0.12 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr6_+_111580508 | 0.12 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr16_+_67563250 | 0.12 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr11_-_61658853 | 0.12 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr10_+_80828774 | 0.12 |
ENST00000334512.5
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr11_-_65429891 | 0.12 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr22_-_39239987 | 0.12 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.2 | 0.6 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.5 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.3 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.2 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.1 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.1 | 0.2 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.4 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.6 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.4 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.7 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:1903273 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 0.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.4 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.2 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) linoleoyl-CoA desaturase activity(GO:0016213) 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |