Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
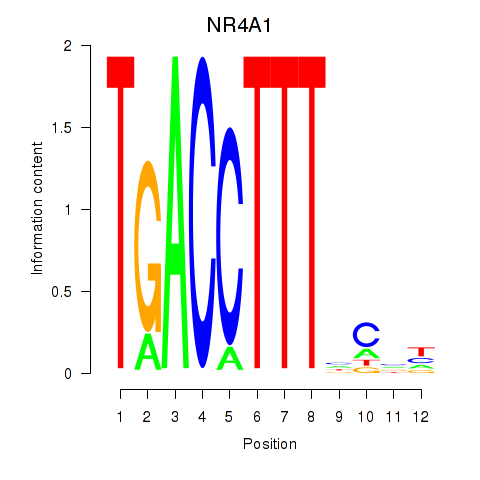
Results for NR4A1
Z-value: 0.26
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.15 | nuclear receptor subfamily 4 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A1 | hg19_v2_chr12_+_52450298_52450388 | -0.99 | 1.1e-02 | Click! |
Activity profile of NR4A1 motif
Sorted Z-values of NR4A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_32476072 | 0.15 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr14_+_21566980 | 0.12 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr17_+_73629500 | 0.11 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr8_-_92053042 | 0.10 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chr3_-_117716418 | 0.10 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr14_+_35591509 | 0.09 |
ENST00000604073.1
|
KIAA0391
|
KIAA0391 |
| chrX_-_108868390 | 0.07 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr16_+_53241854 | 0.07 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_+_19536083 | 0.07 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr2_+_162016827 | 0.06 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_-_72936608 | 0.05 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr12_+_50794947 | 0.05 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr14_-_104387888 | 0.05 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr17_-_66287257 | 0.05 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr17_+_9745786 | 0.05 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr14_+_35591020 | 0.05 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr5_-_87564620 | 0.05 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr12_-_95467267 | 0.05 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr2_+_207024306 | 0.05 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr14_-_101295407 | 0.05 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr4_+_57253672 | 0.04 |
ENST00000602927.1
|
RP11-646I6.5
|
RP11-646I6.5 |
| chr7_-_72936531 | 0.04 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr10_+_26986582 | 0.04 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr4_-_89744365 | 0.04 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr5_-_74162605 | 0.04 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr7_-_37488547 | 0.04 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr4_-_74088800 | 0.04 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr1_-_111743285 | 0.04 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr2_-_25194476 | 0.04 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr11_-_46638378 | 0.04 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr9_+_36572851 | 0.04 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr12_-_109219937 | 0.04 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr17_-_7123021 | 0.04 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr19_+_49713991 | 0.04 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr6_+_133561725 | 0.04 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr10_-_33625154 | 0.04 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr17_-_66287310 | 0.04 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr14_-_95623607 | 0.04 |
ENST00000531162.1
ENST00000529720.1 ENST00000343455.3 |
DICER1
|
dicer 1, ribonuclease type III |
| chr3_+_14989186 | 0.04 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_178257401 | 0.04 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr5_-_126409159 | 0.03 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr8_-_92053212 | 0.03 |
ENST00000285419.3
|
TMEM55A
|
transmembrane protein 55A |
| chr2_+_27282134 | 0.03 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr2_+_162016804 | 0.03 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_55354822 | 0.03 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr14_-_35591679 | 0.03 |
ENST00000557278.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr11_+_7618413 | 0.03 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_32655110 | 0.03 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_+_32654965 | 0.03 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr15_-_42783303 | 0.03 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr12_+_50794891 | 0.03 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr14_+_69951457 | 0.03 |
ENST00000322564.7
|
PLEKHD1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr8_+_133787586 | 0.03 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr7_-_105517021 | 0.03 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr6_+_149068464 | 0.03 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr11_-_113577014 | 0.03 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr3_-_113464906 | 0.03 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr16_+_81272287 | 0.03 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr7_-_99774945 | 0.03 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr10_-_128359074 | 0.03 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr2_+_162016916 | 0.03 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr12_+_21679220 | 0.03 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr12_+_32655048 | 0.03 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_26568965 | 0.03 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr10_-_75634219 | 0.03 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr18_-_60986613 | 0.03 |
ENST00000444484.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr6_+_64346386 | 0.03 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chr6_+_31895480 | 0.03 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr14_+_101295638 | 0.03 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr12_+_96252706 | 0.03 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr2_+_178257372 | 0.03 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr2_+_234216454 | 0.03 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr4_-_89744314 | 0.03 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr11_-_113577052 | 0.02 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr14_-_104387831 | 0.02 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr5_-_142784101 | 0.02 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_+_31895467 | 0.02 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr14_-_95624227 | 0.02 |
ENST00000526495.1
|
DICER1
|
dicer 1, ribonuclease type III |
| chr5_+_159895275 | 0.02 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr1_+_236849754 | 0.02 |
ENST00000542672.1
ENST00000366578.4 |
ACTN2
|
actinin, alpha 2 |
| chr7_-_229557 | 0.02 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr10_+_12110963 | 0.02 |
ENST00000263035.4
ENST00000437298.1 |
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr12_+_54378849 | 0.02 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr7_-_6048702 | 0.02 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr3_-_133648656 | 0.02 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36 |
| chr5_-_53115506 | 0.02 |
ENST00000511953.1
ENST00000504552.1 |
CTD-2081C10.1
|
CTD-2081C10.1 |
| chr11_+_86749035 | 0.02 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr1_+_66820058 | 0.02 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_+_138340049 | 0.02 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr9_-_75567962 | 0.02 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr5_+_35852797 | 0.02 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr10_-_75634326 | 0.02 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr11_+_278365 | 0.02 |
ENST00000534750.1
|
NLRP6
|
NLR family, pyrin domain containing 6 |
| chr1_+_212738676 | 0.02 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr9_+_1051481 | 0.02 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr17_-_38928414 | 0.02 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr7_-_6048650 | 0.02 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr11_+_86748863 | 0.02 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr5_+_73109339 | 0.02 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr17_+_7123125 | 0.02 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr1_+_2398876 | 0.01 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2 |
| chr6_-_130536774 | 0.01 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr1_+_61869748 | 0.01 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr4_-_40632605 | 0.01 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chrX_-_48823165 | 0.01 |
ENST00000419374.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr2_-_44223138 | 0.01 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr11_-_111649015 | 0.01 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_-_142784003 | 0.01 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_29322437 | 0.01 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr16_+_21312170 | 0.01 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr11_+_86748957 | 0.01 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr6_-_152958521 | 0.01 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr3_+_113465866 | 0.01 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr12_+_58335360 | 0.01 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr3_-_44519131 | 0.01 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr12_+_54378923 | 0.01 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr20_-_42815733 | 0.01 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr22_-_31364187 | 0.01 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr10_-_75634260 | 0.01 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr10_+_101542462 | 0.01 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr17_+_41150793 | 0.01 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr15_-_41120896 | 0.01 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr12_+_109577202 | 0.01 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr2_-_207024233 | 0.01 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr10_-_75351088 | 0.01 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr5_-_142784888 | 0.01 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chrX_-_54824673 | 0.01 |
ENST00000218436.6
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr3_+_184037466 | 0.01 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chrX_+_114874727 | 0.01 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr12_+_49687425 | 0.01 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr13_-_30881134 | 0.01 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr7_-_81399355 | 0.01 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_+_35591735 | 0.01 |
ENST00000604948.1
ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391
|
KIAA0391 |
| chr4_+_176986978 | 0.01 |
ENST00000508596.1
ENST00000393643.2 |
WDR17
|
WD repeat domain 17 |
| chr1_+_176432298 | 0.00 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr5_+_127039075 | 0.00 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr4_-_65275162 | 0.00 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr2_-_207024134 | 0.00 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr11_+_120971882 | 0.00 |
ENST00000392793.1
|
TECTA
|
tectorin alpha |
| chr5_-_27038683 | 0.00 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr17_-_36499693 | 0.00 |
ENST00000342292.4
|
GPR179
|
G protein-coupled receptor 179 |
| chr4_-_57253587 | 0.00 |
ENST00000513376.1
ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr4_+_159593271 | 0.00 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr7_+_6048856 | 0.00 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr19_+_13134772 | 0.00 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr8_+_123793633 | 0.00 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr4_-_89744457 | 0.00 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr14_-_24711470 | 0.00 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr18_-_60986962 | 0.00 |
ENST00000333681.4
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr13_-_33112956 | 0.00 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr3_+_14989076 | 0.00 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_-_197036364 | 0.00 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr17_+_41150290 | 0.00 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chrX_+_2746850 | 0.00 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr3_+_101443476 | 0.00 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr3_+_51741072 | 0.00 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr20_-_44485835 | 0.00 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr7_+_99933730 | 0.00 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |