Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
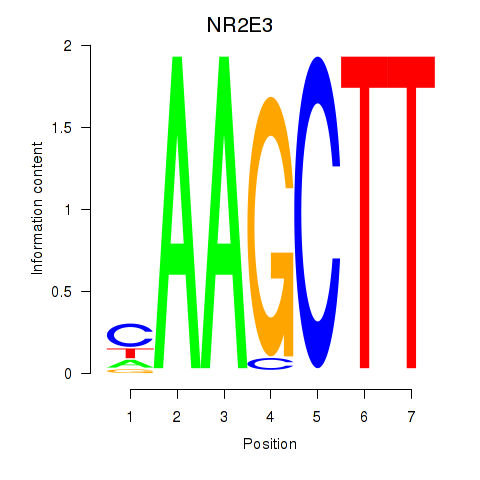
Results for NR2E3
Z-value: 0.81
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_195310802 | 0.42 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr15_+_96897466 | 0.41 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr18_+_55712915 | 0.38 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_+_12008986 | 0.28 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr12_+_32655110 | 0.25 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_121493717 | 0.25 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr19_-_52307357 | 0.23 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr15_+_30375158 | 0.22 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr5_+_59783941 | 0.20 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_-_204183071 | 0.19 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr14_+_71165292 | 0.19 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr12_-_66317967 | 0.19 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr5_-_65018834 | 0.18 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr19_-_14628645 | 0.18 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr2_-_175711924 | 0.18 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr6_-_101329157 | 0.18 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr2_-_177684007 | 0.17 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr5_-_102455801 | 0.17 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr11_+_17298342 | 0.16 |
ENST00000530964.1
|
NUCB2
|
nucleobindin 2 |
| chr9_+_95909309 | 0.15 |
ENST00000366188.2
|
RP11-370F5.4
|
RP11-370F5.4 |
| chr11_+_1891380 | 0.15 |
ENST00000429923.1
ENST00000418975.1 ENST00000406638.2 |
LSP1
|
lymphocyte-specific protein 1 |
| chr2_-_65593784 | 0.15 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_124569864 | 0.15 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr13_+_76378357 | 0.15 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr5_-_59783882 | 0.15 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_-_14596140 | 0.15 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr11_-_32452357 | 0.15 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr5_+_96079240 | 0.15 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr1_+_61547405 | 0.15 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr16_-_75529273 | 0.15 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr10_-_21186144 | 0.14 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_+_32655048 | 0.14 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_-_127840021 | 0.14 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chrX_+_149867681 | 0.14 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr17_-_36413133 | 0.14 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr5_+_179135246 | 0.14 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr13_+_76378407 | 0.13 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr12_+_510795 | 0.13 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr15_-_32695396 | 0.13 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr19_+_37808831 | 0.13 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr2_-_175711978 | 0.13 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr6_-_76203345 | 0.13 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_196159268 | 0.13 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr15_+_79165222 | 0.13 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr12_+_510742 | 0.13 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr1_+_84629976 | 0.13 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_76555657 | 0.13 |
ENST00000307465.4
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr12_-_8765446 | 0.12 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr2_+_7073174 | 0.12 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr17_-_36358166 | 0.12 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr11_+_121447469 | 0.12 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr19_+_42037433 | 0.12 |
ENST00000599316.1
ENST00000599770.1 |
AC006129.1
|
AC006129.1 |
| chr6_+_126221034 | 0.12 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr5_+_75904918 | 0.12 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr15_+_30427352 | 0.12 |
ENST00000569052.1
|
GOLGA8T
|
golgin A8 family, member T |
| chr11_-_71639446 | 0.12 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr4_-_6711558 | 0.11 |
ENST00000320848.6
|
MRFAP1L1
|
Morf4 family associated protein 1-like 1 |
| chrX_-_109590174 | 0.11 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr13_+_76334795 | 0.11 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr12_+_79258444 | 0.11 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr1_+_114472992 | 0.11 |
ENST00000514621.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_-_65659762 | 0.11 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr8_+_9953061 | 0.11 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr5_+_102455853 | 0.11 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_+_122356488 | 0.11 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr3_-_138312971 | 0.11 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr12_+_79258547 | 0.11 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr6_-_108278456 | 0.11 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr8_+_66955648 | 0.11 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr1_-_217804377 | 0.11 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr11_-_73471655 | 0.10 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr4_+_160188306 | 0.10 |
ENST00000510510.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr4_+_76481258 | 0.10 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr4_+_174089904 | 0.10 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr4_-_170948361 | 0.10 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_-_10305302 | 0.10 |
ENST00000592054.1
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr14_+_73525229 | 0.10 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr4_-_39367949 | 0.10 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr4_+_71859156 | 0.10 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr2_-_3584430 | 0.10 |
ENST00000438482.1
ENST00000422961.1 |
AC108488.4
|
AC108488.4 |
| chr22_-_36013368 | 0.10 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr15_-_30706365 | 0.10 |
ENST00000327271.10
ENST00000544495.1 |
GOLGA8R
|
golgin A8 family, member R |
| chr14_-_92247032 | 0.10 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr15_-_77197620 | 0.10 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr4_+_169633310 | 0.09 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_+_123094672 | 0.09 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr14_-_39639523 | 0.09 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr7_+_110731062 | 0.09 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr6_+_109416684 | 0.09 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr17_+_68165657 | 0.09 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr1_+_45140400 | 0.09 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr9_-_90589402 | 0.09 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr14_+_89060749 | 0.09 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr4_-_170947565 | 0.09 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr13_-_103346854 | 0.09 |
ENST00000267273.6
|
METTL21C
|
methyltransferase like 21C |
| chr12_+_41221975 | 0.09 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr3_+_164924716 | 0.09 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr3_+_99536663 | 0.09 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr1_-_35450897 | 0.09 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr10_-_47181681 | 0.09 |
ENST00000452267.1
|
FAM25B
|
family with sequence similarity 25, member B |
| chr1_-_150780757 | 0.09 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr2_-_197664366 | 0.09 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr13_+_76334567 | 0.09 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr1_+_200638629 | 0.09 |
ENST00000568695.1
|
RP11-92G12.3
|
RP11-92G12.3 |
| chr19_-_40596767 | 0.09 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr6_+_12007897 | 0.09 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr8_+_74903580 | 0.09 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr20_-_40247133 | 0.09 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr14_+_58754751 | 0.09 |
ENST00000598233.1
|
AL132989.1
|
AL132989.1 |
| chr20_-_20033052 | 0.09 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr1_+_62439037 | 0.09 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr20_+_43538692 | 0.09 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr4_-_25865159 | 0.09 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr4_+_170581213 | 0.08 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr18_-_66382289 | 0.08 |
ENST00000443099.2
ENST00000562706.1 ENST00000544714.2 |
TMX3
|
thioredoxin-related transmembrane protein 3 |
| chr11_+_65479702 | 0.08 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr2_-_178128250 | 0.08 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr14_-_75083313 | 0.08 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chrX_-_102565858 | 0.08 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr21_+_40752378 | 0.08 |
ENST00000398753.1
ENST00000442773.1 |
WRB
|
tryptophan rich basic protein |
| chr5_+_180682720 | 0.08 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr9_-_116102530 | 0.08 |
ENST00000374195.3
ENST00000341761.4 |
WDR31
|
WD repeat domain 31 |
| chr1_+_45140360 | 0.08 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr5_+_180467259 | 0.08 |
ENST00000515271.1
|
BTNL9
|
butyrophilin-like 9 |
| chr3_+_184033551 | 0.08 |
ENST00000456033.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr2_-_178128149 | 0.08 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr18_-_28622774 | 0.08 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr21_-_16374688 | 0.08 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr18_-_33047039 | 0.08 |
ENST00000591141.1
ENST00000586741.1 |
RP11-322E11.5
|
RP11-322E11.5 |
| chr11_+_17298297 | 0.08 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr15_+_57211318 | 0.08 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr11_-_122929699 | 0.08 |
ENST00000526686.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr13_-_48575376 | 0.08 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr6_-_101329191 | 0.08 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr8_+_38677850 | 0.08 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_-_102962874 | 0.08 |
ENST00000531543.1
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chrX_+_123094369 | 0.08 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr5_-_150473127 | 0.08 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr15_-_33360342 | 0.08 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr10_-_14372870 | 0.08 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr15_-_32747835 | 0.08 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chr11_+_74204883 | 0.08 |
ENST00000528481.1
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr14_-_77542485 | 0.08 |
ENST00000556781.1
ENST00000557526.1 ENST00000555512.1 |
RP11-7F17.3
|
RP11-7F17.3 |
| chr11_+_17298522 | 0.08 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr1_+_28099683 | 0.08 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chrX_-_80065146 | 0.08 |
ENST00000373275.4
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr4_+_174089951 | 0.07 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr10_-_104866395 | 0.07 |
ENST00000458345.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr7_-_79082867 | 0.07 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_-_15149828 | 0.07 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr9_-_112260531 | 0.07 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr18_-_28622699 | 0.07 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr14_-_34420259 | 0.07 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr4_+_71588372 | 0.07 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr7_+_92158083 | 0.07 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr4_+_128554081 | 0.07 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr2_+_102456277 | 0.07 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr1_-_43282906 | 0.07 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr5_-_75008244 | 0.07 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr3_+_57875711 | 0.07 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr6_-_42418999 | 0.07 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr1_-_29557383 | 0.07 |
ENST00000373791.3
ENST00000263702.6 |
MECR
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr18_+_23806437 | 0.07 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr5_+_140710061 | 0.07 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chrX_-_48858630 | 0.07 |
ENST00000376425.3
ENST00000376444.3 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr6_-_29648887 | 0.07 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr2_+_162087577 | 0.07 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_-_16738451 | 0.07 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr10_-_375422 | 0.07 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr1_+_61330931 | 0.07 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr7_-_130080818 | 0.07 |
ENST00000343969.5
ENST00000541543.1 ENST00000489512.1 |
CEP41
|
centrosomal protein 41kDa |
| chr3_-_15374033 | 0.07 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chrX_+_17755563 | 0.07 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr15_+_42787452 | 0.07 |
ENST00000249647.3
|
SNAP23
|
synaptosomal-associated protein, 23kDa |
| chr6_-_4135693 | 0.07 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chrX_+_49832231 | 0.07 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr6_+_54173227 | 0.07 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr6_-_131291572 | 0.07 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_+_31494672 | 0.07 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr13_+_98086445 | 0.07 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr1_-_155904161 | 0.07 |
ENST00000368319.3
|
KIAA0907
|
KIAA0907 |
| chr16_+_50776021 | 0.07 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr5_+_68788594 | 0.07 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr6_+_4776580 | 0.07 |
ENST00000397588.3
|
CDYL
|
chromodomain protein, Y-like |
| chrX_-_48858667 | 0.06 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr19_+_3762703 | 0.06 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr11_-_33913708 | 0.06 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr12_+_96588279 | 0.06 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr15_+_67841330 | 0.06 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr22_+_38382163 | 0.06 |
ENST00000333418.4
ENST00000427034.1 |
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr9_-_104198042 | 0.06 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr4_+_159727272 | 0.06 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr11_-_8795787 | 0.06 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr20_+_34020827 | 0.06 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr1_+_114473350 | 0.06 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr3_-_65583561 | 0.06 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr13_+_76334498 | 0.06 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr9_-_90589586 | 0.06 |
ENST00000325303.8
ENST00000375883.3 |
CDK20
|
cyclin-dependent kinase 20 |
| chr3_-_189839467 | 0.06 |
ENST00000426003.1
|
LEPREL1
|
leprecan-like 1 |
| chr17_-_46691990 | 0.06 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0043353 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:2000282 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.0 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.0 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |