Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
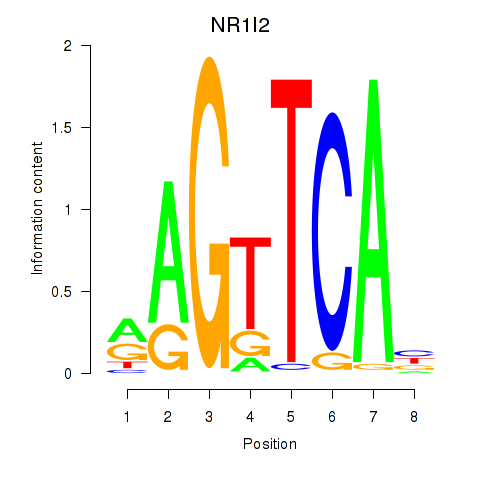
Results for NR1I2
Z-value: 0.50
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.12 | nuclear receptor subfamily 1 group I member 2 |
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_66624527 | 0.31 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr6_-_154751629 | 0.29 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr16_+_691792 | 0.29 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr2_+_233734994 | 0.23 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr6_+_27782788 | 0.21 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr6_+_31515337 | 0.20 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr1_+_11796177 | 0.19 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr1_+_149858461 | 0.18 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr1_+_11796126 | 0.17 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr7_-_150777874 | 0.17 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr6_-_27782548 | 0.17 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr11_-_67141640 | 0.17 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr6_+_26273144 | 0.17 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr19_+_36236491 | 0.16 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr1_+_16083098 | 0.16 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr7_-_150777920 | 0.16 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_+_36236514 | 0.16 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr22_+_50354104 | 0.15 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr1_-_149814478 | 0.15 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr1_+_201979743 | 0.14 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr16_+_84209738 | 0.14 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr16_+_30675654 | 0.14 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr11_-_63993690 | 0.14 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr7_-_150777949 | 0.14 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_-_3547305 | 0.14 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_-_120243545 | 0.14 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr8_-_33370607 | 0.14 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr3_+_112929850 | 0.14 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr16_+_1832902 | 0.14 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr1_+_16083154 | 0.13 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_+_67207838 | 0.13 |
ENST00000566871.1
ENST00000268605.7 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr14_-_65769392 | 0.13 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr17_+_7255208 | 0.12 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr11_+_64879317 | 0.12 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr20_+_62492566 | 0.12 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr1_-_12908578 | 0.12 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr11_-_72414430 | 0.12 |
ENST00000452383.2
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_-_159094194 | 0.12 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr22_+_30821732 | 0.12 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr5_+_177027101 | 0.12 |
ENST00000029410.5
ENST00000510761.1 ENST00000505468.1 |
B4GALT7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 |
| chr17_-_4852332 | 0.12 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chrX_-_30327495 | 0.12 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr2_+_223916862 | 0.12 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr1_+_74701062 | 0.12 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr14_-_95236551 | 0.11 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr1_-_153521714 | 0.11 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr14_+_23340822 | 0.11 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr11_-_72414256 | 0.11 |
ENST00000427971.2
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr9_-_139981121 | 0.11 |
ENST00000596585.1
|
AL807752.1
|
Uncharacterized protein |
| chr16_+_83986827 | 0.10 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr22_+_30821784 | 0.10 |
ENST00000407550.3
|
MTFP1
|
mitochondrial fission process 1 |
| chr10_-_76995675 | 0.10 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr15_+_76030311 | 0.10 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr7_-_56174161 | 0.10 |
ENST00000395422.3
|
CHCHD2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr19_+_39616410 | 0.10 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chrX_+_152990302 | 0.10 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr10_-_103347883 | 0.10 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr19_-_38806540 | 0.10 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr5_+_139055021 | 0.10 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr12_+_34175398 | 0.10 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr3_-_9994021 | 0.10 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr16_+_67207872 | 0.10 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr16_+_28875126 | 0.10 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr19_-_39322497 | 0.10 |
ENST00000221418.4
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr11_+_62649158 | 0.10 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_-_118122996 | 0.10 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr17_+_38497640 | 0.09 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr14_+_73706308 | 0.09 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr16_-_28937027 | 0.09 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr2_+_65663812 | 0.09 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr8_+_99956662 | 0.09 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr8_-_145550337 | 0.09 |
ENST00000531896.1
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr22_+_25615489 | 0.09 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr6_+_106988986 | 0.09 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr7_+_143080063 | 0.09 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr2_+_30454390 | 0.09 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr19_-_38806560 | 0.09 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr3_-_49722523 | 0.09 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr17_-_46682321 | 0.09 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr1_+_45274154 | 0.09 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr5_+_175298573 | 0.09 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr6_-_34664612 | 0.09 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr16_+_619931 | 0.09 |
ENST00000321878.5
ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chrX_-_153192211 | 0.09 |
ENST00000461052.1
ENST00000422091.1 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr19_+_1248547 | 0.08 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr18_+_3466248 | 0.08 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr1_-_229569834 | 0.08 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr9_+_125796806 | 0.08 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr20_+_43343886 | 0.08 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr1_+_223101757 | 0.08 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr11_+_62648336 | 0.08 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr22_+_45098067 | 0.08 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr5_+_139055055 | 0.08 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr11_-_1782625 | 0.08 |
ENST00000438213.1
|
CTSD
|
cathepsin D |
| chr16_-_68002456 | 0.08 |
ENST00000576616.1
ENST00000572037.1 ENST00000338335.3 ENST00000422611.2 ENST00000316341.3 |
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr22_+_29469012 | 0.08 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr17_-_73761222 | 0.08 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr11_-_407103 | 0.08 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr2_-_176046391 | 0.08 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr12_+_6494285 | 0.07 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr5_-_158757895 | 0.07 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr11_+_62439126 | 0.07 |
ENST00000377953.3
|
C11orf83
|
chromosome 11 open reading frame 83 |
| chr19_-_38878632 | 0.07 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr3_+_14474178 | 0.07 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr16_+_28874860 | 0.07 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr17_+_43239191 | 0.07 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr14_+_51026844 | 0.07 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr1_+_43291220 | 0.07 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr12_+_38710555 | 0.07 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr17_-_2169425 | 0.07 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr7_+_97910962 | 0.07 |
ENST00000539286.1
|
BRI3
|
brain protein I3 |
| chr16_-_2770216 | 0.07 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr19_-_38806390 | 0.07 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr2_-_208489275 | 0.07 |
ENST00000272839.3
ENST00000426075.1 |
METTL21A
|
methyltransferase like 21A |
| chr8_+_39972170 | 0.07 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr11_+_67798363 | 0.07 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr19_+_40697514 | 0.07 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr3_+_8543533 | 0.07 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr10_+_80828774 | 0.07 |
ENST00000334512.5
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr8_-_121824374 | 0.07 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr16_-_67281413 | 0.07 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr15_-_94614049 | 0.07 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr7_+_150065879 | 0.07 |
ENST00000397281.2
ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr5_+_175298674 | 0.07 |
ENST00000514150.1
|
CPLX2
|
complexin 2 |
| chr12_-_89746264 | 0.06 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr11_+_63993738 | 0.06 |
ENST00000441250.2
ENST00000279206.3 |
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr18_-_52989525 | 0.06 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr20_-_43438912 | 0.06 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr12_-_49582978 | 0.06 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr8_+_145149930 | 0.06 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr1_+_145209092 | 0.06 |
ENST00000362074.6
ENST00000344859.3 |
NOTCH2NL
|
notch 2 N-terminal like |
| chr13_-_95364389 | 0.06 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr3_+_8543561 | 0.06 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr17_-_7145106 | 0.06 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr1_+_15479054 | 0.06 |
ENST00000376014.3
ENST00000451326.2 |
TMEM51
|
transmembrane protein 51 |
| chr15_-_65067773 | 0.06 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
| chr1_+_196743912 | 0.06 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr1_+_16083123 | 0.06 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr10_+_99185917 | 0.06 |
ENST00000334828.5
|
PGAM1
|
phosphoglycerate mutase 1 (brain) |
| chr15_-_43212836 | 0.06 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr16_+_29991673 | 0.06 |
ENST00000416441.2
|
TAOK2
|
TAO kinase 2 |
| chr17_-_7167279 | 0.06 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr12_-_94673956 | 0.06 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr17_-_56296580 | 0.06 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr9_-_127358087 | 0.06 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr5_-_133747589 | 0.06 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr1_+_36348790 | 0.06 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr5_+_180650271 | 0.05 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr17_+_43239231 | 0.05 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr3_+_112930306 | 0.05 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr7_-_37026108 | 0.05 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_+_99956759 | 0.05 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr9_-_34620440 | 0.05 |
ENST00000421919.1
ENST00000378911.3 ENST00000477738.2 ENST00000341694.2 ENST00000259632.7 ENST00000378913.2 ENST00000378916.4 ENST00000447983.2 |
DCTN3
|
dynactin 3 (p22) |
| chr1_-_157108266 | 0.05 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr9_+_139981513 | 0.05 |
ENST00000535144.1
ENST00000542372.1 |
MAN1B1
|
mannosidase, alpha, class 1B, member 1 |
| chr3_+_8543393 | 0.05 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_-_138665969 | 0.05 |
ENST00000330315.3
|
FOXL2
|
forkhead box L2 |
| chr20_-_3748416 | 0.05 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr1_-_246670614 | 0.05 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr19_+_35485682 | 0.05 |
ENST00000599564.1
|
GRAMD1A
|
GRAM domain containing 1A |
| chr7_-_100844193 | 0.05 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr5_-_158526756 | 0.05 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr18_-_53255766 | 0.05 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr15_+_92397051 | 0.05 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr6_-_107077347 | 0.05 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr4_+_113970772 | 0.05 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr11_-_104480019 | 0.05 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr22_+_25003568 | 0.05 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr20_+_3776371 | 0.05 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr10_-_102890883 | 0.05 |
ENST00000445873.1
|
TLX1NB
|
TLX1 neighbor |
| chr4_+_5712898 | 0.05 |
ENST00000264956.6
ENST00000382674.2 |
EVC
|
Ellis van Creveld syndrome |
| chr12_+_117176090 | 0.05 |
ENST00000257575.4
ENST00000407967.3 ENST00000392549.2 |
RNFT2
|
ring finger protein, transmembrane 2 |
| chr17_+_7254184 | 0.05 |
ENST00000575415.1
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr19_+_45394477 | 0.05 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr17_-_41623716 | 0.05 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr1_-_53387352 | 0.05 |
ENST00000541281.1
|
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr12_+_56511943 | 0.05 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr1_-_173793458 | 0.05 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr16_+_30710462 | 0.05 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr19_+_1438351 | 0.05 |
ENST00000233609.4
|
RPS15
|
ribosomal protein S15 |
| chr15_+_59903975 | 0.05 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr5_-_1801408 | 0.05 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr1_-_55089191 | 0.05 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr22_+_25003626 | 0.05 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr16_-_58328923 | 0.05 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr19_+_11039391 | 0.05 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr22_+_30163340 | 0.05 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr11_+_117073850 | 0.05 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr4_+_22999152 | 0.05 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr15_+_45028719 | 0.05 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr13_-_30424821 | 0.05 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr19_+_1438383 | 0.04 |
ENST00000586686.2
ENST00000591032.1 ENST00000586656.1 ENST00000589656.2 ENST00000593052.1 ENST00000586096.2 ENST00000591804.2 |
RPS15
|
ribosomal protein S15 |
| chr10_+_69865866 | 0.04 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr11_+_308217 | 0.04 |
ENST00000602569.1
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr7_+_97910981 | 0.04 |
ENST00000297290.3
|
BRI3
|
brain protein I3 |
| chr1_+_201979645 | 0.04 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr9_+_139981379 | 0.04 |
ENST00000371589.4
|
MAN1B1
|
mannosidase, alpha, class 1B, member 1 |
| chr12_-_498415 | 0.04 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr17_+_17991197 | 0.04 |
ENST00000225729.3
|
DRG2
|
developmentally regulated GTP binding protein 2 |
| chr2_-_158732340 | 0.04 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.4 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |