Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
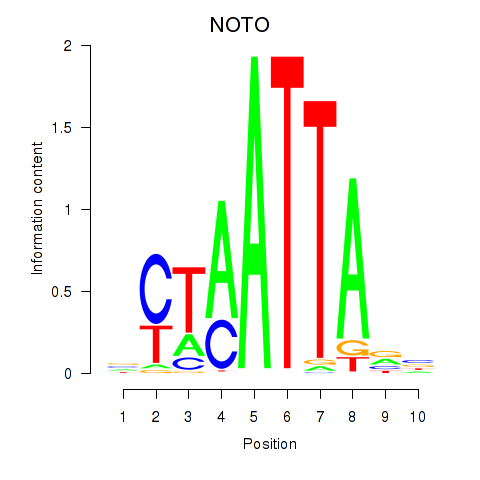
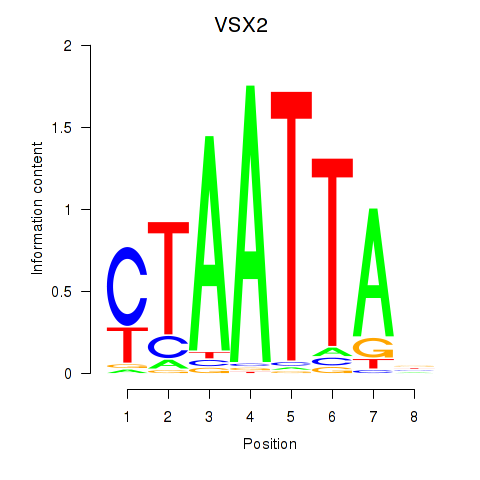
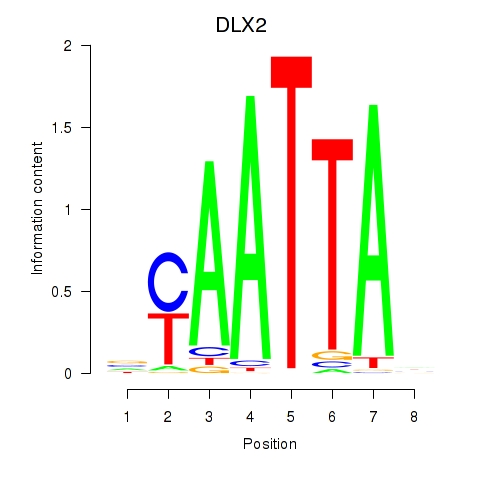
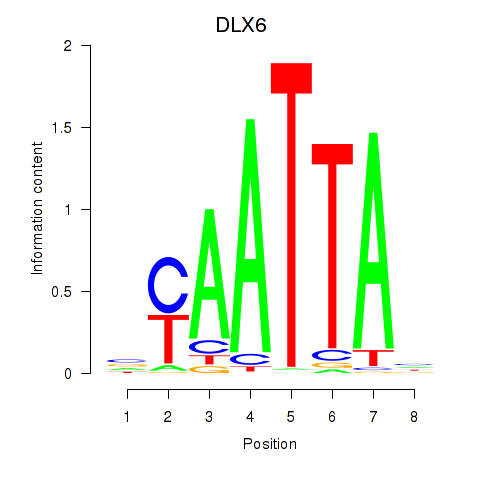
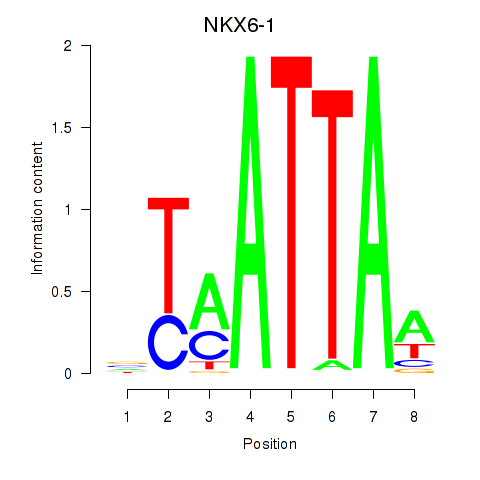
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.56
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NOTO
|
ENSG00000214513.3 | notochord homeobox |
|
VSX2
|
ENSG00000119614.2 | visual system homeobox 2 |
|
DLX2
|
ENSG00000115844.6 | distal-less homeobox 2 |
|
DLX6
|
ENSG00000006377.9 | distal-less homeobox 6 |
|
NKX6-1
|
ENSG00000163623.5 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX6 | hg19_v2_chr7_+_96634850_96634874 | 0.81 | 1.9e-01 | Click! |
| NOTO | hg19_v2_chr2_+_73429386_73429386 | 0.42 | 5.8e-01 | Click! |
| NKX6-1 | hg19_v2_chr4_-_85418103_85418103 | -0.27 | 7.3e-01 | Click! |
| DLX2 | hg19_v2_chr2_-_172967621_172967637 | -0.06 | 9.4e-01 | Click! |
Activity profile of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Sorted Z-values of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_77924627 | 0.47 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr14_-_37051798 | 0.39 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr8_-_25281747 | 0.37 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr6_-_32157947 | 0.32 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr19_-_56110859 | 0.29 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr5_-_126409159 | 0.28 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr8_+_22424551 | 0.25 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr16_+_69345243 | 0.25 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr17_-_71223839 | 0.24 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr5_+_174151536 | 0.22 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr19_+_50016411 | 0.21 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_-_157824292 | 0.21 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr9_-_131486367 | 0.20 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr17_+_4692230 | 0.17 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr11_-_129062093 | 0.17 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr2_-_203735586 | 0.16 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr2_+_175199674 | 0.16 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr1_-_68698222 | 0.15 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr12_+_54378849 | 0.15 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr11_-_8290263 | 0.14 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr16_-_29910853 | 0.14 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr17_+_61151306 | 0.13 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr14_-_75083313 | 0.13 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr9_-_215744 | 0.12 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr9_-_139965000 | 0.12 |
ENST00000409687.3
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr12_-_16761007 | 0.12 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_225600404 | 0.12 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr4_-_89442940 | 0.12 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr16_-_4852915 | 0.12 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr17_-_78450398 | 0.12 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr11_-_62313090 | 0.12 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr19_+_50016610 | 0.12 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_-_55620433 | 0.11 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr7_-_92777606 | 0.11 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr9_-_3469181 | 0.11 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr7_-_84121858 | 0.11 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_43238438 | 0.11 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr15_+_75080883 | 0.11 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr15_-_55562451 | 0.10 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_36020531 | 0.10 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr14_+_38065052 | 0.10 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr14_-_69261310 | 0.10 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_183538319 | 0.10 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr15_+_91416092 | 0.10 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_+_21207503 | 0.10 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_-_183291741 | 0.10 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_140223614 | 0.10 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chrM_+_10053 | 0.09 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr19_+_56111680 | 0.09 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr3_-_120400960 | 0.09 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr1_+_61547894 | 0.09 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr12_-_53994805 | 0.09 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr15_-_52971544 | 0.09 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr4_+_26324474 | 0.08 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_+_74701062 | 0.08 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr16_+_30675654 | 0.08 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr4_-_74486109 | 0.08 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_-_75488984 | 0.08 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr5_+_133562095 | 0.08 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr16_-_30102547 | 0.08 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr2_-_203735484 | 0.08 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr7_-_111424506 | 0.08 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr13_-_36050819 | 0.08 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr10_+_24755416 | 0.08 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr10_-_33623310 | 0.08 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr13_+_110958124 | 0.08 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr5_+_69321074 | 0.08 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr7_-_27179814 | 0.08 |
ENST00000522788.1
ENST00000521779.1 |
HOXA3
|
homeobox A3 |
| chr2_-_87248975 | 0.08 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr1_+_183774240 | 0.08 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr16_+_53133070 | 0.07 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_+_42634844 | 0.07 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr5_+_60933634 | 0.07 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr3_-_114790179 | 0.07 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_-_28621353 | 0.07 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr13_-_110438914 | 0.07 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr14_-_95236551 | 0.07 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr5_+_31193847 | 0.07 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr7_+_73442487 | 0.06 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr20_-_50418947 | 0.06 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr6_-_161695042 | 0.06 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr7_+_99717230 | 0.06 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr16_-_57514277 | 0.06 |
ENST00000562008.1
ENST00000567214.1 |
DOK4
|
docking protein 4 |
| chr16_+_22501658 | 0.06 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_+_24741013 | 0.06 |
ENST00000315183.7
ENST00000395799.3 |
TNRC6A
|
trinucleotide repeat containing 6A |
| chr19_+_48949030 | 0.06 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr14_+_23790655 | 0.06 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr14_-_21994337 | 0.06 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr6_-_31107127 | 0.06 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr13_-_95131923 | 0.06 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr14_-_54425475 | 0.06 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr5_-_141030943 | 0.06 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr5_+_137722255 | 0.06 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr7_-_99716952 | 0.06 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrM_+_10464 | 0.06 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr17_+_73539339 | 0.06 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr12_+_26348582 | 0.06 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr2_-_70780572 | 0.06 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr12_+_49740700 | 0.06 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chrM_+_7586 | 0.05 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr5_+_140855495 | 0.05 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr7_-_104909435 | 0.05 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr16_+_70695570 | 0.05 |
ENST00000597002.1
|
FLJ00418
|
FLJ00418 |
| chr12_-_56694083 | 0.05 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr12_-_118796971 | 0.05 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr6_-_161695074 | 0.05 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr17_-_64225508 | 0.05 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_-_47023455 | 0.05 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr1_-_234667504 | 0.05 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr7_-_111424462 | 0.05 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chrX_+_78003204 | 0.05 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr14_-_57197224 | 0.05 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr2_-_216003127 | 0.05 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr7_-_99716940 | 0.05 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr18_-_68004529 | 0.05 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr14_+_72052983 | 0.05 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr12_-_16760021 | 0.05 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr17_+_73452545 | 0.05 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr4_+_3388057 | 0.05 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr17_+_42264556 | 0.05 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr17_+_46970134 | 0.05 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr1_+_154401791 | 0.05 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr13_-_46716969 | 0.05 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr4_-_74486217 | 0.05 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_14029283 | 0.05 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr8_+_77593448 | 0.05 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chrX_+_10031499 | 0.05 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr1_-_68698197 | 0.04 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chrM_-_14670 | 0.04 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_-_140223670 | 0.04 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr11_-_33913708 | 0.04 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr11_-_96076334 | 0.04 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr6_-_47445214 | 0.04 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr17_-_27418537 | 0.04 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr2_-_190446738 | 0.04 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr7_+_129015484 | 0.04 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr4_+_169418255 | 0.04 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_-_5463175 | 0.04 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr12_-_10978957 | 0.04 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr4_+_66536248 | 0.04 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr12_-_28125638 | 0.04 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr15_+_44092784 | 0.04 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr2_-_70780770 | 0.04 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr12_-_118797475 | 0.04 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr4_+_183065793 | 0.04 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr7_-_73038867 | 0.04 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr6_+_114178512 | 0.04 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr20_-_50418972 | 0.04 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr17_+_46970127 | 0.04 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr18_+_61445007 | 0.04 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr20_-_42939782 | 0.04 |
ENST00000396825.3
|
FITM2
|
fat storage-inducing transmembrane protein 2 |
| chr12_-_28123206 | 0.04 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr10_+_4828815 | 0.04 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr10_-_21806759 | 0.04 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr7_-_20826504 | 0.04 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr3_+_141105705 | 0.04 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_+_210443993 | 0.04 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr4_-_138453606 | 0.04 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr13_+_109248500 | 0.04 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr16_-_69390209 | 0.04 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr16_+_89334512 | 0.04 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chrX_-_133792480 | 0.04 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr18_+_61445205 | 0.04 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr16_-_28621298 | 0.04 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr20_-_50419055 | 0.04 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_+_188664988 | 0.04 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr13_-_31040060 | 0.04 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr1_+_36621529 | 0.03 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr8_-_42358742 | 0.03 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr6_-_32908792 | 0.03 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr5_+_140529630 | 0.03 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr7_-_83278322 | 0.03 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr18_-_53089723 | 0.03 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr9_+_12775011 | 0.03 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr1_+_183774285 | 0.03 |
ENST00000539189.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr1_-_21625486 | 0.03 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr2_+_190541153 | 0.03 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr1_+_28586006 | 0.03 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr15_+_91411810 | 0.03 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr11_-_126870655 | 0.03 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr3_-_11623804 | 0.03 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_+_61548225 | 0.03 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr4_-_177116772 | 0.03 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr14_-_38064198 | 0.03 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr10_+_111765562 | 0.03 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr2_+_234826016 | 0.03 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr8_-_74495065 | 0.03 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr22_+_46449674 | 0.03 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr1_+_168250194 | 0.03 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chrX_-_73512358 | 0.03 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr11_+_110001723 | 0.03 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr18_+_32173276 | 0.03 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr22_+_44427230 | 0.03 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr17_+_48823975 | 0.03 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr7_-_111032971 | 0.03 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr3_-_141747950 | 0.03 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chrX_+_100224676 | 0.03 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr17_+_47448102 | 0.03 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of NOTO_VSX2_DLX2_DLX6_NKX6-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.0 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.0 | GO:2001190 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.0 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |