Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
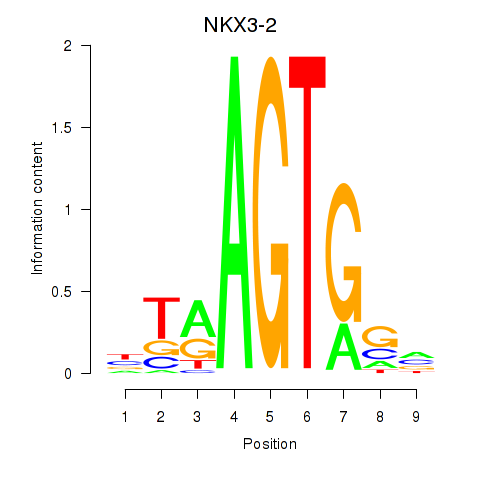
Results for NKX3-2
Z-value: 0.64
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.7 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | 0.37 | 6.3e-01 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_149402009 | 0.31 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr19_+_18544045 | 0.23 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr19_+_54609230 | 0.22 |
ENST00000420296.1
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr1_-_204183071 | 0.22 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr17_+_34171081 | 0.22 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr14_+_21566980 | 0.21 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr3_-_42003613 | 0.21 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr7_-_128415844 | 0.21 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr1_-_235098935 | 0.20 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr4_-_85771168 | 0.20 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chrY_+_15418467 | 0.19 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr2_+_232572361 | 0.19 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr6_+_155443048 | 0.19 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_-_150039170 | 0.18 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr14_+_64565442 | 0.17 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr6_+_149539053 | 0.16 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr12_-_54978086 | 0.16 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr11_+_77532233 | 0.16 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr16_+_69333585 | 0.16 |
ENST00000570054.2
|
RP11-343C2.11
|
Uncharacterized protein |
| chr5_-_59481406 | 0.15 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_-_1590418 | 0.15 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr7_-_50628745 | 0.15 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr7_-_8276508 | 0.15 |
ENST00000401396.1
ENST00000317367.5 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr7_-_22862406 | 0.15 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr14_-_107283278 | 0.14 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr19_-_56048456 | 0.14 |
ENST00000413299.1
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr6_-_131299929 | 0.14 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_98444386 | 0.14 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr12_+_50794947 | 0.14 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr1_+_202431859 | 0.14 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_75904918 | 0.13 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chrX_+_38420783 | 0.13 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr1_-_1208851 | 0.13 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr11_-_123065989 | 0.13 |
ENST00000448775.2
|
CLMP
|
CXADR-like membrane protein |
| chr17_+_38083977 | 0.13 |
ENST00000578802.1
ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4
|
RP11-387H17.4 |
| chr13_+_44596471 | 0.13 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chr9_-_139268068 | 0.12 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr12_-_21927736 | 0.12 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr12_-_10022735 | 0.12 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr9_+_35906176 | 0.11 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr7_-_45957011 | 0.11 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr12_+_14518598 | 0.11 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr14_+_105927191 | 0.11 |
ENST00000550551.1
|
MTA1
|
metastasis associated 1 |
| chr8_+_87111059 | 0.11 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr7_-_45956856 | 0.11 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr7_-_120498357 | 0.11 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr15_-_100258029 | 0.11 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr3_-_107777208 | 0.11 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr20_+_42984330 | 0.11 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr1_-_226065330 | 0.11 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr19_+_7694623 | 0.11 |
ENST00000594797.1
ENST00000456958.3 ENST00000601406.1 |
PET100
|
PET100 homolog (S. cerevisiae) |
| chr3_+_169629354 | 0.11 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr19_+_36266417 | 0.11 |
ENST00000378944.5
ENST00000007510.4 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr1_-_21059029 | 0.11 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr11_-_8739383 | 0.10 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr6_-_32784687 | 0.10 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr2_-_27294500 | 0.10 |
ENST00000447619.1
ENST00000429985.1 ENST00000456793.1 |
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
| chr2_+_102927962 | 0.10 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr19_-_6393216 | 0.10 |
ENST00000595047.1
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chr20_+_33462745 | 0.09 |
ENST00000488172.1
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr11_-_119993734 | 0.09 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr7_+_99971129 | 0.09 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr14_+_78227105 | 0.09 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chrX_+_48660565 | 0.09 |
ENST00000413163.2
ENST00000441703.1 ENST00000426196.1 |
HDAC6
|
histone deacetylase 6 |
| chr2_-_70780770 | 0.09 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr1_+_205473720 | 0.09 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr5_+_118406796 | 0.09 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr2_-_45166338 | 0.09 |
ENST00000437916.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr10_+_98064085 | 0.09 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr6_-_138833630 | 0.09 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chrX_+_38420623 | 0.09 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr1_-_33642151 | 0.09 |
ENST00000543586.1
|
TRIM62
|
tripartite motif containing 62 |
| chr3_-_69249863 | 0.09 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr11_-_72852958 | 0.08 |
ENST00000458644.2
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr12_+_56075330 | 0.08 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr1_-_78444738 | 0.08 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr12_-_113658892 | 0.08 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr15_-_40401062 | 0.08 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr11_-_77531752 | 0.08 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr8_-_144674284 | 0.08 |
ENST00000528519.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr8_+_22414182 | 0.08 |
ENST00000524057.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr5_+_118604439 | 0.08 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr10_+_7745303 | 0.08 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr14_+_58894404 | 0.08 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr3_+_184056614 | 0.08 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr7_+_110731062 | 0.08 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr14_+_31494672 | 0.08 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_-_62432641 | 0.08 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr1_-_24438664 | 0.08 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chr2_-_24270217 | 0.08 |
ENST00000295148.4
ENST00000406895.3 |
C2orf44
|
chromosome 2 open reading frame 44 |
| chr1_-_222886526 | 0.08 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr11_-_93474645 | 0.08 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr12_-_122712038 | 0.07 |
ENST00000413918.1
ENST00000443649.3 |
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr2_+_29336855 | 0.07 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr12_+_96337061 | 0.07 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr1_+_206579736 | 0.07 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr5_+_78407602 | 0.07 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr10_-_27529779 | 0.07 |
ENST00000426079.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_-_122247879 | 0.07 |
ENST00000545861.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr4_+_159727272 | 0.07 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr4_-_11430221 | 0.07 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr8_-_27469383 | 0.07 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr5_-_149792295 | 0.07 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr5_+_64859066 | 0.07 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr3_-_116163830 | 0.07 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr10_-_7708918 | 0.07 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr6_+_56820152 | 0.07 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr5_+_75904950 | 0.07 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr15_+_42694573 | 0.07 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr6_+_24357131 | 0.07 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr19_+_57999079 | 0.07 |
ENST00000426954.2
ENST00000354197.4 ENST00000523882.1 ENST00000520540.1 ENST00000519310.1 ENST00000442920.2 ENST00000523312.1 ENST00000424930.2 |
ZNF419
|
zinc finger protein 419 |
| chr9_-_34397800 | 0.07 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr8_-_29592736 | 0.07 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr12_+_50794891 | 0.07 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr10_-_27529486 | 0.07 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr14_-_62162541 | 0.07 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr17_+_58755184 | 0.07 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr19_-_11266437 | 0.06 |
ENST00000586708.1
ENST00000591396.1 ENST00000592967.1 ENST00000585486.1 ENST00000585567.1 |
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr7_-_117512264 | 0.06 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr10_-_29923893 | 0.06 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr2_+_233385173 | 0.06 |
ENST00000449534.2
|
PRSS56
|
protease, serine, 56 |
| chr5_-_137514333 | 0.06 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr16_-_3350614 | 0.06 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr19_+_16999654 | 0.06 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr7_-_111424462 | 0.06 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_+_136289030 | 0.06 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr17_-_40168698 | 0.06 |
ENST00000590774.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr7_-_111424506 | 0.06 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr17_+_53828432 | 0.06 |
ENST00000573500.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr1_-_48866517 | 0.06 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr4_+_26324474 | 0.06 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_32687221 | 0.06 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_-_137514288 | 0.06 |
ENST00000454473.1
ENST00000418329.1 ENST00000455658.2 ENST00000230901.5 ENST00000402931.1 |
BRD8
|
bromodomain containing 8 |
| chr2_+_173940163 | 0.06 |
ENST00000539448.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr1_+_119957554 | 0.06 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr11_-_77531858 | 0.06 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr6_+_18155560 | 0.06 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr16_-_20566616 | 0.06 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr19_+_52074502 | 0.06 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr7_-_56160625 | 0.06 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr19_-_10628098 | 0.06 |
ENST00000590601.1
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr1_+_31769836 | 0.06 |
ENST00000344147.5
ENST00000373714.1 ENST00000546109.1 ENST00000422613.2 |
ZCCHC17
|
zinc finger, CCHC domain containing 17 |
| chr6_-_30080876 | 0.06 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr12_-_42983478 | 0.06 |
ENST00000345127.3
ENST00000547113.1 |
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr18_+_21464737 | 0.05 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr12_-_95397442 | 0.05 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr7_+_155090271 | 0.05 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr3_+_125694347 | 0.05 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr9_+_5890802 | 0.05 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr21_-_36259445 | 0.05 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr3_-_160117301 | 0.05 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr9_+_112542591 | 0.05 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr1_-_152086556 | 0.05 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr7_+_99971068 | 0.05 |
ENST00000198536.2
ENST00000453419.1 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chrX_+_100333709 | 0.05 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr14_+_31494841 | 0.05 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr10_-_47173994 | 0.05 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr7_-_102119342 | 0.05 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr10_-_131762105 | 0.05 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr12_+_57522692 | 0.05 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr17_-_42295870 | 0.05 |
ENST00000526094.1
ENST00000529383.1 ENST00000530828.1 |
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr6_+_159084188 | 0.05 |
ENST00000367081.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr5_-_77072085 | 0.05 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr7_-_56160666 | 0.05 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr3_-_160116995 | 0.05 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr3_-_149688502 | 0.05 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr1_-_35497506 | 0.05 |
ENST00000317538.5
ENST00000373340.2 ENST00000357182.4 |
ZMYM6
|
zinc finger, MYM-type 6 |
| chr5_+_118604385 | 0.05 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr3_-_130746462 | 0.05 |
ENST00000505545.1
|
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr1_-_154164534 | 0.05 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr5_+_112074029 | 0.05 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr21_+_30671690 | 0.05 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr1_-_156710916 | 0.05 |
ENST00000368211.4
|
MRPL24
|
mitochondrial ribosomal protein L24 |
| chr5_+_179135246 | 0.05 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr2_-_25016251 | 0.05 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr3_-_52008016 | 0.05 |
ENST00000489595.2
ENST00000461108.1 ENST00000395008.2 ENST00000525795.1 ENST00000361143.5 |
RP11-155D18.14
ABHD14B
|
Poly(rC)-binding protein 4 abhydrolase domain containing 14B |
| chr4_+_148402069 | 0.05 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chr16_+_3194211 | 0.05 |
ENST00000428155.1
|
CASP16
|
caspase 16, apoptosis-related cysteine peptidase (putative) |
| chr2_-_182545603 | 0.05 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr14_+_58894706 | 0.05 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr15_-_43559055 | 0.05 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr4_-_48018680 | 0.04 |
ENST00000513178.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr10_-_75571566 | 0.04 |
ENST00000299641.4
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr22_-_41636929 | 0.04 |
ENST00000216241.9
|
CHADL
|
chondroadherin-like |
| chr18_-_6414884 | 0.04 |
ENST00000317931.7
ENST00000284898.6 ENST00000400104.3 |
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr1_+_161195835 | 0.04 |
ENST00000545897.1
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr2_-_190044480 | 0.04 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr10_+_16478942 | 0.04 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr8_-_101734170 | 0.04 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_122711968 | 0.04 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr3_+_50192537 | 0.04 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_-_160117035 | 0.04 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr17_-_74137374 | 0.04 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr15_-_37393406 | 0.04 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr6_+_31082603 | 0.04 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr11_-_106889157 | 0.04 |
ENST00000282249.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chr3_+_48481658 | 0.04 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr14_-_50778931 | 0.04 |
ENST00000555423.1
ENST00000421284.3 |
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase |
| chr14_+_21156915 | 0.04 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr9_+_15553055 | 0.04 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.0 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.0 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |