Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
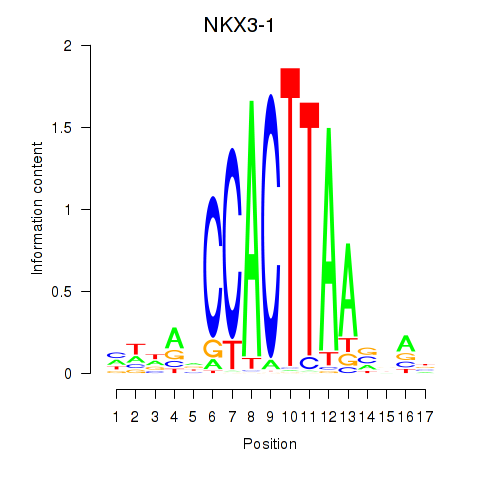
Results for NKX3-1
Z-value: 0.99
Transcription factors associated with NKX3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-1
|
ENSG00000167034.9 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-1 | hg19_v2_chr8_-_23540402_23540466 | 0.99 | 1.1e-02 | Click! |
Activity profile of NKX3-1 motif
Sorted Z-values of NKX3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_149785236 | 2.47 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr11_-_9482010 | 0.97 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr16_+_3068393 | 0.86 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr5_+_176873446 | 0.71 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr19_-_45457264 | 0.70 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chr6_-_154751629 | 0.49 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr19_+_50433476 | 0.49 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr11_-_64684672 | 0.48 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr11_-_62323702 | 0.44 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr16_+_1832902 | 0.43 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr11_+_114168085 | 0.37 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr6_-_26216872 | 0.36 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr11_+_65266507 | 0.36 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_+_110011571 | 0.36 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr16_-_31076332 | 0.32 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr21_-_46964306 | 0.32 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr17_+_42422629 | 0.32 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chr16_-_31076273 | 0.30 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr6_+_26020672 | 0.30 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr19_+_56111680 | 0.29 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr19_-_55791431 | 0.29 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr10_-_103347883 | 0.29 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr12_+_70219052 | 0.28 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr15_-_93353028 | 0.28 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr17_+_67498396 | 0.28 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_-_74726283 | 0.27 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chrX_+_153237740 | 0.27 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr1_-_156399184 | 0.27 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr11_+_67798090 | 0.27 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr19_+_17830051 | 0.27 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr14_-_65409502 | 0.26 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr20_-_2821271 | 0.26 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr16_+_28985542 | 0.26 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr17_+_42422662 | 0.26 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr19_-_55791540 | 0.25 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr3_-_49726104 | 0.25 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr16_+_32264040 | 0.25 |
ENST00000398664.3
|
TP53TG3D
|
TP53 target 3D |
| chr7_+_141463897 | 0.25 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr1_+_218683438 | 0.24 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr11_+_67798114 | 0.24 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr4_-_90756769 | 0.24 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr16_+_73420942 | 0.24 |
ENST00000554640.1
ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1
|
RP11-140I24.1 |
| chr5_+_150404904 | 0.24 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_-_1685718 | 0.23 |
ENST00000472884.2
ENST00000489363.1 ENST00000308132.6 ENST00000463238.1 |
FAM53A
|
family with sequence similarity 53, member A |
| chr14_+_97925151 | 0.23 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr8_+_27169138 | 0.23 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr19_-_56110859 | 0.23 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr15_-_31521567 | 0.23 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr11_-_118901559 | 0.22 |
ENST00000330775.7
ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr17_-_39661849 | 0.22 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr20_-_2821756 | 0.22 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr16_-_3074231 | 0.21 |
ENST00000572355.1
ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr3_-_10334617 | 0.21 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr16_-_69373396 | 0.21 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr10_+_46994087 | 0.21 |
ENST00000374317.1
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr11_+_64073022 | 0.21 |
ENST00000406310.1
ENST00000000442.6 ENST00000539594.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr9_+_134065506 | 0.21 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr6_-_26124138 | 0.21 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr19_-_2944907 | 0.21 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr19_-_1592652 | 0.21 |
ENST00000156825.1
ENST00000434436.3 |
MBD3
|
methyl-CpG binding domain protein 3 |
| chr3_+_98482175 | 0.21 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr3_-_48632593 | 0.20 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr11_+_94277017 | 0.20 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr10_+_18629628 | 0.19 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr17_-_7307358 | 0.19 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_173686303 | 0.19 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_67798363 | 0.19 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chrX_-_153236819 | 0.19 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr5_-_43412418 | 0.19 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr16_+_83986827 | 0.18 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr2_-_27545921 | 0.18 |
ENST00000402310.1
ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr19_+_52704346 | 0.18 |
ENST00000462990.1
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr14_-_54420133 | 0.18 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr5_-_1799965 | 0.18 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr14_-_74551172 | 0.18 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr19_-_42894420 | 0.18 |
ENST00000597255.1
ENST00000222032.5 |
CNFN
|
cornifelin |
| chr3_-_178976996 | 0.18 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr19_+_10362882 | 0.18 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr17_+_42422637 | 0.17 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr4_-_105416039 | 0.17 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr6_-_83775489 | 0.17 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr17_+_67498295 | 0.17 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr10_+_135204338 | 0.17 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr3_+_113251143 | 0.17 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr19_+_16308711 | 0.17 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr17_+_7358889 | 0.17 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr11_+_65337901 | 0.16 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr11_-_117688216 | 0.16 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr11_+_64685026 | 0.16 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr10_-_28571015 | 0.16 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_57039739 | 0.16 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr4_+_159131596 | 0.15 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr14_-_68066975 | 0.15 |
ENST00000559581.1
ENST00000560722.1 ENST00000559415.1 ENST00000216452.4 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr22_+_44427230 | 0.15 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr2_+_238877424 | 0.15 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr9_+_134065519 | 0.15 |
ENST00000531600.1
|
NUP214
|
nucleoporin 214kDa |
| chr1_+_151372010 | 0.15 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr6_+_43603552 | 0.15 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr5_-_176889381 | 0.15 |
ENST00000393563.4
ENST00000512501.1 |
DBN1
|
drebrin 1 |
| chr7_+_142374104 | 0.14 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr17_-_7145475 | 0.14 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr12_-_118628315 | 0.14 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr11_-_104480019 | 0.14 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr5_+_112312416 | 0.14 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr14_-_65409438 | 0.14 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr6_+_37321823 | 0.14 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr16_+_56969284 | 0.14 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr3_-_15106747 | 0.14 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chrX_-_153237258 | 0.14 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr6_-_31648150 | 0.13 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr1_+_27114589 | 0.13 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr1_+_186798073 | 0.13 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr3_+_52489503 | 0.13 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr17_-_39661947 | 0.13 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr8_+_30300119 | 0.13 |
ENST00000520191.1
|
RBPMS
|
RNA binding protein with multiple splicing |
| chr10_-_61900762 | 0.13 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_-_15544099 | 0.13 |
ENST00000599910.2
|
WIZ
|
widely interspaced zinc finger motifs |
| chr1_-_173793458 | 0.13 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr1_-_110155671 | 0.13 |
ENST00000351050.3
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr7_+_16700806 | 0.12 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr7_-_7680601 | 0.12 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr3_+_39509070 | 0.12 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr19_+_13080296 | 0.12 |
ENST00000317060.2
|
DAND5
|
DAN domain family member 5, BMP antagonist |
| chr19_+_17186577 | 0.12 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr6_-_116833500 | 0.12 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr1_-_153931052 | 0.12 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr3_+_39509163 | 0.12 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr14_+_95078714 | 0.12 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr19_-_44174330 | 0.12 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_+_161475208 | 0.11 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr13_+_24144509 | 0.11 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_77325545 | 0.11 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chr6_+_108616054 | 0.11 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr8_+_128426535 | 0.11 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr6_-_144385698 | 0.11 |
ENST00000444202.1
ENST00000437412.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr19_+_17970677 | 0.11 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr10_-_120938303 | 0.10 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr12_-_125398602 | 0.10 |
ENST00000541272.1
ENST00000535131.1 |
UBC
|
ubiquitin C |
| chr12_-_49333446 | 0.10 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr12_-_53298841 | 0.10 |
ENST00000293308.6
|
KRT8
|
keratin 8 |
| chr19_+_17970693 | 0.10 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr17_-_76870222 | 0.09 |
ENST00000585421.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr19_-_11266471 | 0.09 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr9_+_116263639 | 0.09 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr19_-_55791563 | 0.09 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr20_-_36661826 | 0.09 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr16_+_89628778 | 0.09 |
ENST00000472354.1
|
RPL13
|
ribosomal protein L13 |
| chr22_+_18632666 | 0.08 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr11_+_101983176 | 0.08 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr6_-_144416737 | 0.08 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr18_+_61445007 | 0.08 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr20_-_45318051 | 0.08 |
ENST00000372102.3
|
TP53RK
|
TP53 regulating kinase |
| chr11_+_57435441 | 0.08 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr3_-_39196049 | 0.08 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr17_+_41150479 | 0.08 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chrY_+_15418467 | 0.08 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr5_+_125758813 | 0.08 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr20_-_50722183 | 0.08 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr19_+_16308659 | 0.08 |
ENST00000590263.1
ENST00000590756.1 ENST00000541844.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr2_-_36779411 | 0.08 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr7_-_112430647 | 0.08 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr14_-_47812321 | 0.08 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr1_-_39339777 | 0.08 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr7_-_6066183 | 0.08 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chrX_-_107681633 | 0.08 |
ENST00000394872.2
ENST00000334504.7 |
COL4A6
|
collagen, type IV, alpha 6 |
| chr4_-_90757364 | 0.08 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_+_125758865 | 0.07 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr6_-_33285505 | 0.07 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr12_+_96883347 | 0.07 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr19_-_44174305 | 0.07 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr19_-_5719860 | 0.07 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr12_+_6602517 | 0.07 |
ENST00000315579.5
ENST00000539714.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chr17_-_295730 | 0.07 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr5_+_96211643 | 0.07 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr14_+_23654525 | 0.07 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr19_+_37341260 | 0.07 |
ENST00000589046.1
|
ZNF345
|
zinc finger protein 345 |
| chr8_+_77593448 | 0.06 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr19_-_14889349 | 0.06 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr18_+_61445205 | 0.06 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr16_-_69760409 | 0.06 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr7_-_2883928 | 0.06 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr9_+_74729511 | 0.06 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr3_-_156840776 | 0.06 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr11_+_86106208 | 0.06 |
ENST00000528728.1
|
CCDC81
|
coiled-coil domain containing 81 |
| chr4_+_15376165 | 0.05 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_32169920 | 0.05 |
ENST00000373672.3
ENST00000373668.3 |
COL16A1
|
collagen, type XVI, alpha 1 |
| chr19_-_42931567 | 0.05 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr22_-_50523760 | 0.05 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_-_22063787 | 0.05 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_173793641 | 0.05 |
ENST00000361951.4
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr3_-_18480260 | 0.05 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chrX_+_9502971 | 0.05 |
ENST00000452824.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr11_-_26588634 | 0.05 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr17_-_76870126 | 0.05 |
ENST00000586057.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr7_+_100303676 | 0.05 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr6_+_26124373 | 0.05 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr1_+_173793777 | 0.05 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr5_-_139937895 | 0.05 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr19_+_58144529 | 0.04 |
ENST00000347302.3
ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211
|
zinc finger protein 211 |
| chr7_+_33168856 | 0.04 |
ENST00000432983.1
|
BBS9
|
Bardet-Biedl syndrome 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.3 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.3 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.2 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:1904344 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.2 | GO:0048392 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) mesodermal cell fate determination(GO:0007500) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.4 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |