Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
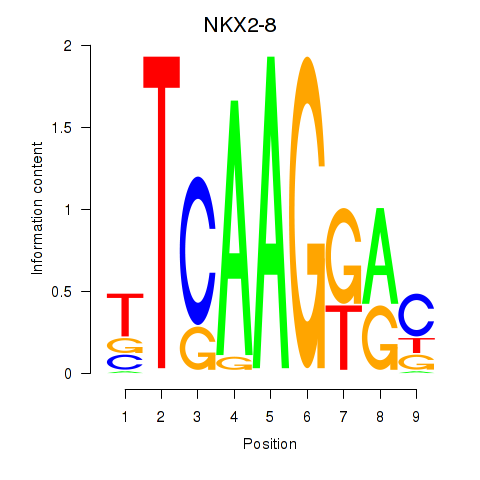
Results for NKX2-8
Z-value: 0.45
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | 0.82 | 1.8e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_25451065 | 0.28 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr17_+_73455788 | 0.28 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr17_+_76210367 | 0.27 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr1_+_180165672 | 0.27 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr6_-_32145861 | 0.22 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr15_-_75248954 | 0.22 |
ENST00000499788.2
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr11_-_67141640 | 0.22 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr16_+_88493879 | 0.19 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr19_+_2249308 | 0.18 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr1_+_154401791 | 0.18 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr22_-_21213676 | 0.18 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr7_-_99716914 | 0.15 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_+_59705928 | 0.15 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr9_+_131873659 | 0.15 |
ENST00000452489.2
ENST00000347048.4 ENST00000357197.4 ENST00000445241.1 ENST00000355007.3 ENST00000414331.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_+_100054606 | 0.15 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr7_+_150929550 | 0.14 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr8_-_144679264 | 0.14 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr7_-_156803329 | 0.14 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr16_-_75301886 | 0.14 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_+_35222629 | 0.14 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr17_+_46184911 | 0.13 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr16_+_28875268 | 0.13 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr2_-_36779411 | 0.13 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr20_+_55967129 | 0.12 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr19_-_18391708 | 0.12 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr2_-_132249955 | 0.12 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr16_+_28875126 | 0.12 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr17_+_34901353 | 0.11 |
ENST00000593016.1
|
GGNBP2
|
gametogenetin binding protein 2 |
| chr1_-_68698222 | 0.11 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr18_+_3450161 | 0.11 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_+_49658855 | 0.11 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr2_-_197675000 | 0.11 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr7_+_99816859 | 0.11 |
ENST00000317271.2
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing |
| chr1_+_16083098 | 0.10 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr3_-_114790179 | 0.10 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr17_-_202579 | 0.10 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr12_-_49504623 | 0.10 |
ENST00000550137.1
ENST00000547382.1 |
LMBR1L
|
limb development membrane protein 1-like |
| chr11_+_64808675 | 0.10 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr17_+_46185111 | 0.10 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr11_-_117170403 | 0.10 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr2_+_62132800 | 0.09 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr22_+_38609538 | 0.09 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr19_-_39322497 | 0.09 |
ENST00000221418.4
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr3_-_47950745 | 0.09 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr12_-_49504655 | 0.09 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr4_-_164253738 | 0.09 |
ENST00000509586.1
ENST00000504391.1 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chrM_+_8366 | 0.09 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr21_+_33671264 | 0.09 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr9_+_109685630 | 0.09 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chrX_+_48681768 | 0.09 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr1_+_16083123 | 0.09 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_+_73472575 | 0.08 |
ENST00000375248.5
|
KIAA0195
|
KIAA0195 |
| chr8_-_61429315 | 0.08 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chr22_+_29834572 | 0.08 |
ENST00000354373.2
|
RFPL1
|
ret finger protein-like 1 |
| chr12_+_13349711 | 0.08 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr19_-_39322299 | 0.08 |
ENST00000601094.1
ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr6_+_30844192 | 0.08 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_-_17622269 | 0.08 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr1_+_74701062 | 0.08 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr5_+_35852797 | 0.08 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr1_-_155006224 | 0.08 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr14_+_92980111 | 0.08 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr12_+_50505762 | 0.07 |
ENST00000550487.1
ENST00000317943.2 |
COX14
|
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr19_+_36486078 | 0.07 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chr3_-_53290016 | 0.07 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr19_+_34856078 | 0.07 |
ENST00000589399.1
ENST00000589640.1 ENST00000591204.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr10_+_115674530 | 0.07 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr17_+_8191815 | 0.07 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr1_+_179050512 | 0.07 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr11_-_133826852 | 0.07 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr19_-_3600549 | 0.07 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr5_+_92919043 | 0.07 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr8_+_27182862 | 0.07 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr8_-_145018905 | 0.06 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr14_-_88282595 | 0.06 |
ENST00000554519.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr8_+_128426535 | 0.06 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chrX_-_109590174 | 0.06 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr16_+_28874860 | 0.06 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr12_+_121416489 | 0.06 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr17_+_7591639 | 0.06 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr10_+_74653330 | 0.06 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr7_-_766879 | 0.06 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr13_+_88325498 | 0.06 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr18_-_72920372 | 0.05 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr12_-_102591604 | 0.05 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr3_+_126423045 | 0.05 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chrX_-_64196307 | 0.05 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_-_99057551 | 0.05 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr22_+_20877924 | 0.05 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr16_-_30441293 | 0.05 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr4_+_15341442 | 0.05 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr3_+_47021168 | 0.05 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr6_-_32806506 | 0.05 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr17_+_7591747 | 0.05 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr12_-_49504449 | 0.05 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr2_-_133429091 | 0.05 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr6_-_31830655 | 0.05 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chrX_-_48776292 | 0.05 |
ENST00000376509.4
|
PIM2
|
pim-2 oncogene |
| chr2_+_101591314 | 0.05 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr12_+_66218212 | 0.05 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr3_-_178103144 | 0.05 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr17_+_76210267 | 0.05 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr5_+_149737202 | 0.05 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr1_+_113010056 | 0.04 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr10_-_15762124 | 0.04 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr8_+_67341239 | 0.04 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr1_+_43291220 | 0.04 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr3_-_121740969 | 0.04 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr17_+_39994032 | 0.04 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr15_-_70387120 | 0.04 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr1_+_55464600 | 0.04 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr7_-_86974767 | 0.04 |
ENST00000610086.1
|
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chr21_+_33671160 | 0.04 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_-_87039662 | 0.04 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr4_-_153601136 | 0.04 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr5_-_78281775 | 0.04 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr6_-_32731299 | 0.04 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr8_+_67405794 | 0.04 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr22_+_39101728 | 0.04 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr12_-_118406777 | 0.03 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr9_+_21409146 | 0.03 |
ENST00000380205.1
|
IFNA8
|
interferon, alpha 8 |
| chr22_+_38453207 | 0.03 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr17_-_46703826 | 0.03 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr12_-_12837423 | 0.03 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr12_+_13349650 | 0.03 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr2_-_68547019 | 0.03 |
ENST00000409862.1
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr1_-_95007193 | 0.03 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr17_+_73642486 | 0.03 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr22_-_50523760 | 0.03 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr7_-_92855762 | 0.03 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr13_+_108921977 | 0.03 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr1_+_76251912 | 0.03 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr3_-_180397256 | 0.03 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr1_-_27998689 | 0.03 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr3_-_187455680 | 0.03 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr19_-_9003586 | 0.03 |
ENST00000380951.5
|
MUC16
|
mucin 16, cell surface associated |
| chr18_+_44497455 | 0.02 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr19_-_11347173 | 0.02 |
ENST00000587656.1
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr6_+_41010293 | 0.02 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr17_+_62503073 | 0.02 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chrX_-_110655306 | 0.02 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr3_+_101498026 | 0.02 |
ENST00000495842.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr11_-_104827425 | 0.02 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_18417948 | 0.02 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr13_-_46756351 | 0.02 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_+_125524785 | 0.02 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr2_+_11752379 | 0.02 |
ENST00000396123.1
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_-_178984759 | 0.02 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr15_+_51973680 | 0.02 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr19_+_34856141 | 0.02 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr17_-_8055747 | 0.02 |
ENST00000317276.4
ENST00000581703.1 |
PER1
|
period circadian clock 1 |
| chr3_+_177545563 | 0.02 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr20_-_48747662 | 0.02 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr20_-_43936937 | 0.02 |
ENST00000342716.4
ENST00000353917.5 ENST00000360607.6 ENST00000372751.4 |
MATN4
|
matrilin 4 |
| chr20_-_44485835 | 0.02 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr15_+_93749295 | 0.02 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chrX_+_41548220 | 0.02 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr17_+_61086917 | 0.02 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_+_591524 | 0.02 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr1_+_226013047 | 0.02 |
ENST00000366837.4
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr10_-_118032979 | 0.02 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr3_-_8811288 | 0.02 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr4_+_184826418 | 0.02 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr18_-_13915530 | 0.02 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr16_+_8807419 | 0.02 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chrX_-_53461288 | 0.02 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chrX_-_72095808 | 0.02 |
ENST00000373529.5
|
DMRTC1
|
DMRT-like family C1 |
| chr3_+_137728842 | 0.02 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr17_-_8198636 | 0.02 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr14_-_107219365 | 0.02 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr5_-_179107975 | 0.02 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr2_-_68547061 | 0.01 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr1_-_94147385 | 0.01 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr15_-_60683326 | 0.01 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr15_-_28419569 | 0.01 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr22_+_31518938 | 0.01 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr12_-_46663734 | 0.01 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr14_-_58893876 | 0.01 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_-_114343039 | 0.01 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_177502438 | 0.01 |
ENST00000443670.1
|
AC017048.4
|
long intergenic non-protein coding RNA 1117 |
| chr15_-_50558223 | 0.01 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr14_-_36990354 | 0.01 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr5_+_140474181 | 0.01 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr8_+_104310661 | 0.01 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr1_-_104238912 | 0.01 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr1_+_12040238 | 0.01 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr12_-_10320190 | 0.01 |
ENST00000543993.1
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr22_-_50523807 | 0.01 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr8_+_23430157 | 0.01 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chr3_-_139195350 | 0.01 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr1_+_173793777 | 0.01 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr3_+_187930719 | 0.01 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_+_43602750 | 0.00 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr12_+_66218598 | 0.00 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chrX_+_118533269 | 0.00 |
ENST00000336249.7
|
SLC25A43
|
solute carrier family 25, member 43 |
| chr4_-_110723134 | 0.00 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr4_-_70080449 | 0.00 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr10_-_135171178 | 0.00 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr1_+_41157361 | 0.00 |
ENST00000427410.2
ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr14_+_58894404 | 0.00 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
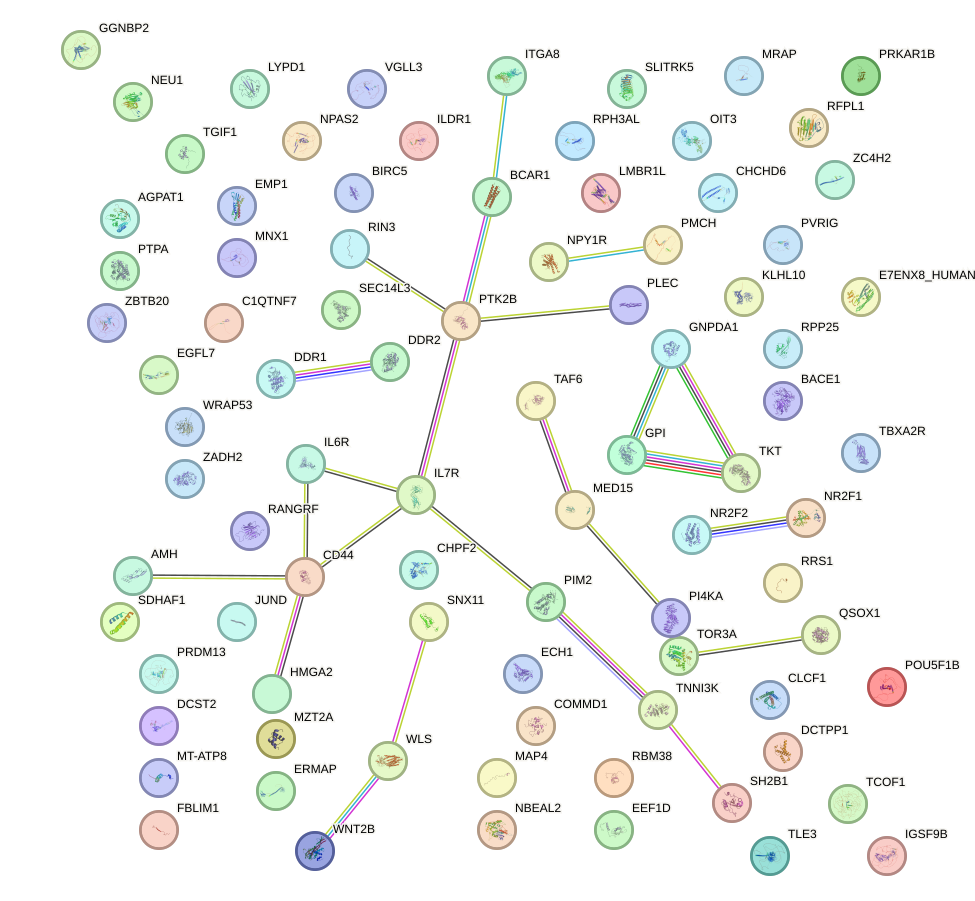
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |