Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for NKX2-3
Z-value: 1.13
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-3 | hg19_v2_chr10_+_101292684_101292706 | -0.98 | 2.1e-02 | Click! |
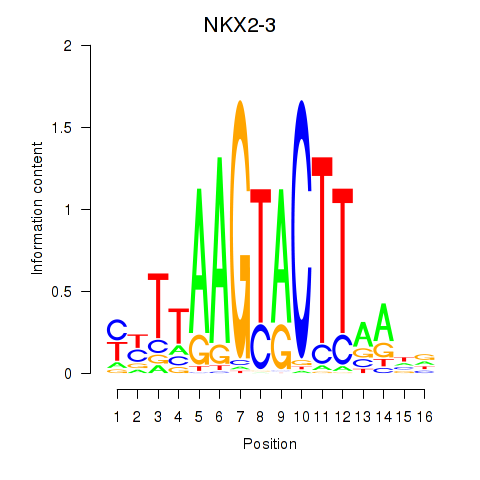
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26027480 | 0.98 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr6_+_42123141 | 0.70 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr11_+_65266507 | 0.62 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr17_-_15469590 | 0.58 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr4_-_111536534 | 0.58 |
ENST00000503456.1
ENST00000513690.1 |
RP11-380D23.2
|
RP11-380D23.2 |
| chr4_-_90758227 | 0.55 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_+_109602039 | 0.54 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr19_-_44174330 | 0.49 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr6_-_26216872 | 0.46 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr3_-_167191814 | 0.45 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr1_-_182573514 | 0.41 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr19_+_45409011 | 0.39 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr19_+_13051206 | 0.39 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr15_-_36544450 | 0.38 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr20_-_1317555 | 0.38 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr12_-_102591604 | 0.37 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr10_+_35464513 | 0.37 |
ENST00000494479.1
ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM
|
cAMP responsive element modulator |
| chr3_+_98482175 | 0.36 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr16_+_32264040 | 0.36 |
ENST00000398664.3
|
TP53TG3D
|
TP53 target 3D |
| chr14_+_45366518 | 0.36 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr10_+_129785574 | 0.34 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr16_+_56969284 | 0.33 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr13_+_23993097 | 0.29 |
ENST00000443092.1
|
SACS-AS1
|
SACS antisense RNA 1 |
| chr5_+_140213815 | 0.29 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr3_+_145782358 | 0.28 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr17_-_80797886 | 0.28 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr10_-_36813162 | 0.28 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr7_+_141463897 | 0.27 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr19_-_38743878 | 0.27 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr5_-_159846066 | 0.27 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr4_+_76439649 | 0.27 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr2_-_37501692 | 0.26 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr16_+_8736232 | 0.26 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr2_+_232135245 | 0.25 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr12_-_123201337 | 0.24 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr7_-_2883650 | 0.24 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr5_+_150404904 | 0.23 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr16_+_31225337 | 0.23 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr12_-_104531945 | 0.23 |
ENST00000551446.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr12_+_93963590 | 0.22 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr12_+_111051832 | 0.22 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr3_+_113775594 | 0.22 |
ENST00000479882.1
ENST00000493014.1 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr19_-_33182616 | 0.22 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr20_+_60962143 | 0.22 |
ENST00000343986.4
|
RPS21
|
ribosomal protein S21 |
| chr1_+_225140372 | 0.21 |
ENST00000366848.1
ENST00000439375.2 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr4_-_84035905 | 0.21 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr1_+_35220613 | 0.21 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr12_-_48119301 | 0.21 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr1_-_35325400 | 0.21 |
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12 |
| chr3_-_171527560 | 0.21 |
ENST00000331659.2
|
PP13439
|
PP13439 |
| chr12_+_111051902 | 0.21 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr4_+_80584903 | 0.20 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr4_-_75695366 | 0.20 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr5_-_132200477 | 0.20 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr1_+_43824669 | 0.20 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chr13_-_50018140 | 0.20 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr4_-_140201333 | 0.20 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr3_-_69062764 | 0.19 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_+_23851928 | 0.19 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr6_-_144329531 | 0.19 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr6_-_32160622 | 0.18 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr10_+_1120312 | 0.18 |
ENST00000436154.1
|
WDR37
|
WD repeat domain 37 |
| chr20_+_57427765 | 0.18 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr19_-_46288917 | 0.18 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr3_-_49058479 | 0.18 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr12_+_48178706 | 0.17 |
ENST00000599515.1
|
AC004466.1
|
Uncharacterized protein |
| chr8_+_1919481 | 0.17 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr11_+_35211429 | 0.17 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr8_-_101719159 | 0.17 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_-_39924349 | 0.17 |
ENST00000602153.1
|
RPS16
|
ribosomal protein S16 |
| chr7_-_80548667 | 0.17 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_+_155278625 | 0.17 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr3_-_180397256 | 0.17 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr12_+_123464607 | 0.16 |
ENST00000543566.1
ENST00000315580.5 ENST00000542099.1 ENST00000392435.2 ENST00000413381.2 ENST00000426960.2 ENST00000453766.2 |
ARL6IP4
|
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr2_+_71357744 | 0.16 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr1_+_26147319 | 0.16 |
ENST00000374300.3
|
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr1_-_232598163 | 0.16 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_+_36142147 | 0.16 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr19_+_33865218 | 0.16 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr3_+_23847432 | 0.16 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr20_+_57464200 | 0.16 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr9_+_131447342 | 0.16 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr8_+_91952750 | 0.16 |
ENST00000521366.1
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1 |
| chr2_-_201753980 | 0.16 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr2_-_31440377 | 0.15 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr18_-_74839891 | 0.15 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr4_+_76439665 | 0.15 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr11_+_118889142 | 0.15 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr11_+_118889456 | 0.14 |
ENST00000528230.1
ENST00000525303.1 ENST00000434101.2 ENST00000359005.4 ENST00000533058.1 |
TRAPPC4
|
trafficking protein particle complex 4 |
| chr17_+_53345080 | 0.14 |
ENST00000572002.1
|
HLF
|
hepatic leukemia factor |
| chr1_+_156561533 | 0.14 |
ENST00000368234.3
ENST00000368235.3 ENST00000368233.3 |
APOA1BP
|
apolipoprotein A-I binding protein |
| chr12_-_125398654 | 0.14 |
ENST00000541645.1
ENST00000540351.1 |
UBC
|
ubiquitin C |
| chr1_+_155278539 | 0.14 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr16_+_69458537 | 0.14 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr16_+_2255841 | 0.14 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr4_-_84035868 | 0.14 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr4_+_81118647 | 0.13 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr4_-_39033963 | 0.13 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr2_+_64068844 | 0.13 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr18_+_43684310 | 0.13 |
ENST00000592471.1
ENST00000585518.1 |
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr13_-_44735393 | 0.13 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr20_-_33539655 | 0.12 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr8_+_11961898 | 0.12 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr6_-_116833500 | 0.12 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr5_-_110848189 | 0.12 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr4_+_146403912 | 0.12 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr12_+_71833550 | 0.12 |
ENST00000266674.5
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr16_+_2255710 | 0.12 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr4_+_48343339 | 0.12 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr9_-_25678856 | 0.12 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr12_+_6309963 | 0.11 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr6_-_33239712 | 0.11 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_+_113217043 | 0.11 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr14_+_36295638 | 0.11 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr10_+_18041218 | 0.11 |
ENST00000480516.1
ENST00000457860.1 |
TMEM236
|
TMEM236 |
| chr1_-_155990580 | 0.11 |
ENST00000531917.1
ENST00000480567.1 ENST00000526212.1 ENST00000529008.1 ENST00000496742.1 ENST00000295702.4 |
SSR2
|
signal sequence receptor, beta (translocon-associated protein beta) |
| chr10_+_116697946 | 0.11 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr7_-_105221898 | 0.11 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chrX_-_69509738 | 0.11 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr19_+_39616410 | 0.11 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr1_-_33116128 | 0.11 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr3_+_183967409 | 0.11 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr4_-_104020968 | 0.11 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr19_-_42746714 | 0.11 |
ENST00000222330.3
|
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr6_-_137539651 | 0.11 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chrX_-_108976410 | 0.10 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr12_+_123465033 | 0.10 |
ENST00000454885.2
|
ARL6IP4
|
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr7_+_56032652 | 0.10 |
ENST00000437587.1
|
GBAS
|
glioblastoma amplified sequence |
| chr7_-_2883928 | 0.10 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr8_+_91803695 | 0.10 |
ENST00000417640.2
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1 |
| chr4_-_90757364 | 0.10 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_-_12946236 | 0.10 |
ENST00000589272.1
ENST00000393233.2 |
RTBDN
|
retbindin |
| chr11_-_10822029 | 0.10 |
ENST00000528839.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_174669653 | 0.10 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_144303093 | 0.10 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_+_41157671 | 0.10 |
ENST00000534399.1
ENST00000372653.1 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr11_+_6226782 | 0.10 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr3_-_69062790 | 0.10 |
ENST00000540955.1
ENST00000456376.1 |
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr17_-_76870126 | 0.10 |
ENST00000586057.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr4_-_100212132 | 0.10 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr2_+_202125219 | 0.10 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr6_+_36562132 | 0.10 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr6_-_150212029 | 0.10 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr19_+_3178736 | 0.10 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr12_-_104532062 | 0.10 |
ENST00000240055.3
|
NFYB
|
nuclear transcription factor Y, beta |
| chr19_-_45579762 | 0.10 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr17_+_12569472 | 0.10 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr10_+_17794251 | 0.09 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr6_-_33239612 | 0.09 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_+_174670143 | 0.09 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr16_+_23765948 | 0.09 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr1_+_113217073 | 0.09 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr15_+_69452959 | 0.09 |
ENST00000261858.2
|
GLCE
|
glucuronic acid epimerase |
| chr1_-_26147149 | 0.09 |
ENST00000536896.1
|
AL020996.1
|
Uncharacterized protein |
| chr19_-_49955096 | 0.09 |
ENST00000595550.1
|
PIH1D1
|
PIH1 domain containing 1 |
| chr1_+_109632425 | 0.09 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr2_+_64069459 | 0.09 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr8_+_19674651 | 0.09 |
ENST00000397977.3
|
INTS10
|
integrator complex subunit 10 |
| chrX_-_134049262 | 0.09 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr15_-_101142401 | 0.09 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr3_-_52002403 | 0.09 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr22_+_39101728 | 0.09 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr16_-_18911366 | 0.09 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr16_-_15474904 | 0.09 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr1_-_68962805 | 0.09 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr2_-_36825281 | 0.09 |
ENST00000405912.3
ENST00000379245.4 |
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr12_-_48119340 | 0.08 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chrX_-_108976449 | 0.08 |
ENST00000469857.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr4_-_90758118 | 0.08 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr7_+_7196565 | 0.08 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr6_+_83073334 | 0.08 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr16_-_31076332 | 0.08 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr11_+_94277017 | 0.08 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr18_-_25616519 | 0.08 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr4_-_52883786 | 0.08 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr10_+_103348031 | 0.08 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr17_-_76870222 | 0.08 |
ENST00000585421.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr5_+_49961727 | 0.08 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr9_+_131644398 | 0.08 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr8_-_99954788 | 0.08 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr2_+_132044330 | 0.08 |
ENST00000416266.1
|
CYP4F31P
|
cytochrome P450, family 4, subfamily F, polypeptide 31, pseudogene |
| chr2_+_234621551 | 0.08 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr14_+_100531738 | 0.08 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr14_+_100531615 | 0.08 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr22_+_45714301 | 0.08 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr2_+_64073187 | 0.07 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr14_+_55494323 | 0.07 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr6_-_2971494 | 0.07 |
ENST00000380539.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr10_-_104262460 | 0.07 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr3_-_101039402 | 0.07 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr22_+_32439019 | 0.07 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr18_-_64271316 | 0.07 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr19_-_49955050 | 0.07 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr11_+_7595136 | 0.07 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr14_+_67831576 | 0.07 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr15_+_43477455 | 0.07 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr6_+_83072923 | 0.07 |
ENST00000535040.1
|
TPBG
|
trophoblast glycoprotein |
| chr11_+_12399071 | 0.07 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr1_+_65613340 | 0.07 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr15_-_101142362 | 0.07 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:2000646 | regulation of Cdc42 protein signal transduction(GO:0032489) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.2 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.3 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.2 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.0 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.0 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |