Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
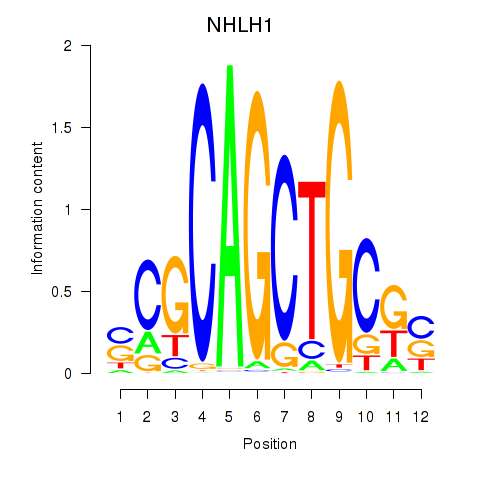
Results for NHLH1
Z-value: 0.66
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | 0.77 | 2.3e-01 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_21905120 | 0.57 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr1_+_38273818 | 0.47 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr1_+_38273988 | 0.34 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr2_-_287320 | 0.32 |
ENST00000401503.1
|
FAM150B
|
family with sequence similarity 150, member B |
| chr16_+_77246337 | 0.32 |
ENST00000563157.1
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr6_+_26156551 | 0.31 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr20_-_62493217 | 0.31 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr5_+_133562095 | 0.30 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr3_-_133614467 | 0.26 |
ENST00000469959.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chrX_-_45710920 | 0.26 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr11_+_22646739 | 0.24 |
ENST00000428556.2
|
AC103801.2
|
AC103801.2 |
| chr9_+_2017063 | 0.23 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_73184588 | 0.23 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr8_+_97506033 | 0.22 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr19_+_39989535 | 0.22 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr2_-_7005785 | 0.21 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr4_+_3768075 | 0.20 |
ENST00000509482.1
ENST00000330055.5 |
ADRA2C
|
adrenoceptor alpha 2C |
| chr2_-_96811170 | 0.20 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr10_-_103347883 | 0.20 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr5_+_176873789 | 0.20 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr15_+_90735145 | 0.18 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr8_-_144897549 | 0.18 |
ENST00000356994.2
ENST00000320476.3 |
SCRIB
|
scribbled planar cell polarity protein |
| chr17_+_73512594 | 0.18 |
ENST00000333213.6
|
TSEN54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr22_-_21984282 | 0.18 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr10_+_102758105 | 0.18 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr11_+_394145 | 0.17 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr4_+_102268904 | 0.17 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr8_-_144897138 | 0.17 |
ENST00000377533.3
|
SCRIB
|
scribbled planar cell polarity protein |
| chr2_+_85981008 | 0.17 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr17_+_40688190 | 0.17 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr13_-_26625169 | 0.17 |
ENST00000319420.3
|
SHISA2
|
shisa family member 2 |
| chr20_-_31172598 | 0.17 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr5_+_176873446 | 0.16 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr1_+_180165672 | 0.16 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_-_133747551 | 0.16 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr2_+_233734994 | 0.16 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr17_+_74261413 | 0.16 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr2_+_191513587 | 0.16 |
ENST00000416973.1
ENST00000426601.1 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr7_+_72848092 | 0.15 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr22_-_24989014 | 0.15 |
ENST00000318753.8
|
FAM211B
|
family with sequence similarity 211, member B |
| chrX_-_47004437 | 0.15 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chrX_+_129305623 | 0.15 |
ENST00000257017.4
|
RAB33A
|
RAB33A, member RAS oncogene family |
| chr20_-_32274179 | 0.15 |
ENST00000343380.5
|
E2F1
|
E2F transcription factor 1 |
| chr14_+_102027688 | 0.15 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr12_+_56137064 | 0.14 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr16_-_67694129 | 0.14 |
ENST00000602320.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr15_-_53082178 | 0.14 |
ENST00000305901.5
|
ONECUT1
|
one cut homeobox 1 |
| chr1_+_113217309 | 0.14 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr6_+_30848829 | 0.14 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr18_+_43914159 | 0.14 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr2_+_11052054 | 0.14 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr12_+_122064673 | 0.13 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr12_+_122064398 | 0.13 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr1_+_20915409 | 0.13 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr16_-_88772761 | 0.13 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr6_+_44095263 | 0.13 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr3_-_178789220 | 0.12 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr11_-_6341844 | 0.12 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr6_-_90121938 | 0.12 |
ENST00000369415.4
|
RRAGD
|
Ras-related GTP binding D |
| chr19_-_55791058 | 0.12 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr3_-_32022733 | 0.12 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr4_+_89300158 | 0.12 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_-_34126730 | 0.12 |
ENST00000361264.4
|
DCAF12
|
DDB1 and CUL4 associated factor 12 |
| chr16_-_67693846 | 0.12 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr19_+_13228917 | 0.12 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr16_-_29910365 | 0.12 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr16_+_771663 | 0.12 |
ENST00000568916.1
|
FAM173A
|
family with sequence similarity 173, member A |
| chr1_-_27693349 | 0.12 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr9_-_140095186 | 0.12 |
ENST00000409012.4
|
TPRN
|
taperin |
| chr13_+_88325498 | 0.12 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr19_+_39989580 | 0.11 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr22_+_45072925 | 0.11 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr1_+_32538492 | 0.11 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr11_-_66725837 | 0.11 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr2_-_239148599 | 0.11 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr16_-_31106211 | 0.11 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr9_-_111929560 | 0.11 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr16_-_70285797 | 0.11 |
ENST00000435634.1
|
EXOSC6
|
exosome component 6 |
| chr6_+_30848557 | 0.11 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr14_-_37051798 | 0.11 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr22_-_42342692 | 0.11 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr17_+_74261277 | 0.11 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr16_-_31106048 | 0.11 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr4_-_2758015 | 0.11 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr15_+_80215477 | 0.11 |
ENST00000542003.1
|
C15ORF37
|
C15orf37 protein; Uncharacterized protein |
| chr5_-_178772424 | 0.11 |
ENST00000251582.7
ENST00000274609.5 |
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr16_+_88772866 | 0.11 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr16_+_3014269 | 0.10 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr19_+_41107249 | 0.10 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr7_+_150065879 | 0.10 |
ENST00000397281.2
ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr19_-_47104118 | 0.10 |
ENST00000593888.1
ENST00000602017.1 |
AC011551.3
PPP5D1
|
Uncharacterized protein PPP5 tetratricopeptide repeat domain containing 1 |
| chr3_-_87040259 | 0.10 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr17_-_31204124 | 0.10 |
ENST00000579584.1
ENST00000318217.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr8_+_128747661 | 0.10 |
ENST00000259523.6
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr20_+_30193083 | 0.10 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr19_+_36132631 | 0.10 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr22_-_20850070 | 0.10 |
ENST00000440659.2
ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22
|
kelch-like family member 22 |
| chr5_+_52776228 | 0.10 |
ENST00000256759.3
|
FST
|
follistatin |
| chr2_-_230135937 | 0.10 |
ENST00000392054.3
ENST00000409462.1 ENST00000392055.3 |
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr20_-_62680984 | 0.10 |
ENST00000340356.7
|
SOX18
|
SRY (sex determining region Y)-box 18 |
| chr19_-_45737469 | 0.10 |
ENST00000413988.1
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr12_+_123459747 | 0.10 |
ENST00000228922.7
ENST00000537966.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr5_+_139027877 | 0.09 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr11_+_65383227 | 0.09 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr15_+_40650408 | 0.09 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr19_-_17366257 | 0.09 |
ENST00000594059.1
|
AC010646.3
|
Uncharacterized protein |
| chr22_+_38054721 | 0.09 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr7_-_74867509 | 0.09 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr1_+_113217345 | 0.09 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr10_+_94833642 | 0.09 |
ENST00000224356.4
ENST00000394139.1 |
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr9_+_140119618 | 0.09 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chr11_-_64014379 | 0.09 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr5_+_52776449 | 0.09 |
ENST00000396947.3
|
FST
|
follistatin |
| chr10_+_12391685 | 0.09 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr11_+_60635035 | 0.09 |
ENST00000278853.5
|
ZP1
|
zona pellucida glycoprotein 1 (sperm receptor) |
| chr17_+_74723031 | 0.09 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr3_-_5229982 | 0.09 |
ENST00000600805.1
|
AC026202.1
|
Uncharacterized protein |
| chr7_+_766320 | 0.09 |
ENST00000297440.6
ENST00000313147.5 |
HEATR2
|
HEAT repeat containing 2 |
| chr6_-_31681839 | 0.09 |
ENST00000409239.1
ENST00000461287.1 |
LY6G6E
XXbac-BPG32J3.20
|
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr11_+_827553 | 0.09 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr3_-_64253655 | 0.09 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr11_-_6341724 | 0.09 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr14_-_54423529 | 0.08 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr2_-_73340146 | 0.08 |
ENST00000258098.6
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I) |
| chr19_-_47734448 | 0.08 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr22_+_45072958 | 0.08 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr4_-_80994471 | 0.08 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr14_-_24911868 | 0.08 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr19_+_12949251 | 0.08 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr12_-_67197760 | 0.08 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr3_-_99833333 | 0.08 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_-_52887034 | 0.08 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr18_+_3448455 | 0.08 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr19_-_3061397 | 0.08 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr6_+_43395272 | 0.08 |
ENST00000372530.4
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr17_+_74722912 | 0.08 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr20_+_35234137 | 0.08 |
ENST00000344795.3
ENST00000373852.5 |
C20orf24
|
chromosome 20 open reading frame 24 |
| chr8_-_38008783 | 0.08 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chrX_-_153285251 | 0.08 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr12_-_54813229 | 0.08 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr11_-_65374430 | 0.08 |
ENST00000532507.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr8_+_128747757 | 0.08 |
ENST00000517291.1
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr3_-_172428842 | 0.07 |
ENST00000424772.1
|
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr16_+_75033210 | 0.07 |
ENST00000566250.1
ENST00000567962.1 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr22_-_50964849 | 0.07 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr20_-_35274548 | 0.07 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr11_-_1606513 | 0.07 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr1_-_11107280 | 0.07 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr12_-_54779511 | 0.07 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr2_+_198570081 | 0.07 |
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr19_-_40730820 | 0.07 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr22_+_45098067 | 0.07 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr19_-_55628927 | 0.07 |
ENST00000263433.3
ENST00000376393.2 |
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C |
| chr12_+_53491220 | 0.07 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr20_+_49575342 | 0.07 |
ENST00000244051.1
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr7_+_766850 | 0.07 |
ENST00000437419.1
ENST00000440747.1 |
HEATR2
|
HEAT repeat containing 2 |
| chr11_+_62554860 | 0.07 |
ENST00000533861.1
ENST00000333449.4 |
TMEM179B
|
transmembrane protein 179B |
| chr15_+_79603404 | 0.07 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chr2_-_72375167 | 0.07 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr4_-_54930790 | 0.07 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr17_-_73761222 | 0.07 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr1_-_38273840 | 0.07 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr15_+_74833518 | 0.07 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr19_+_17858509 | 0.07 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr4_+_89299885 | 0.07 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr5_-_133747589 | 0.07 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr4_-_80994210 | 0.06 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_+_955448 | 0.06 |
ENST00000379370.2
|
AGRN
|
agrin |
| chr14_-_64971893 | 0.06 |
ENST00000555220.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr20_+_49348081 | 0.06 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr11_+_394196 | 0.06 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr16_-_69373396 | 0.06 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr1_-_32687923 | 0.06 |
ENST00000309777.6
ENST00000344461.3 ENST00000373593.1 ENST00000545122.1 |
TMEM234
|
transmembrane protein 234 |
| chr12_+_113796347 | 0.06 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr1_-_3713042 | 0.06 |
ENST00000378251.1
|
LRRC47
|
leucine rich repeat containing 47 |
| chr11_-_61124776 | 0.06 |
ENST00000542361.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr19_+_12780512 | 0.06 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr2_-_73511407 | 0.06 |
ENST00000520530.2
|
FBXO41
|
F-box protein 41 |
| chrX_-_153285395 | 0.06 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr19_-_17445613 | 0.06 |
ENST00000159087.4
|
ANO8
|
anoctamin 8 |
| chr11_-_14913765 | 0.06 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr6_+_159290917 | 0.06 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr7_-_100171270 | 0.06 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr20_-_43438912 | 0.06 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr1_-_6662919 | 0.06 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr17_+_65040678 | 0.06 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr9_+_100615536 | 0.06 |
ENST00000375123.3
|
FOXE1
|
forkhead box E1 (thyroid transcription factor 2) |
| chr9_-_131940526 | 0.06 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr22_-_50964558 | 0.06 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr11_-_31531121 | 0.06 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr17_+_28256874 | 0.06 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr16_+_5121814 | 0.06 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr1_+_32538520 | 0.06 |
ENST00000438825.1
ENST00000456834.2 ENST00000373634.4 ENST00000427288.1 |
TMEM39B
|
transmembrane protein 39B |
| chr5_-_148758839 | 0.06 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr19_-_46272462 | 0.06 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr17_+_81037473 | 0.06 |
ENST00000320095.7
|
METRNL
|
meteorin, glial cell differentiation regulator-like |
| chr17_+_74864476 | 0.06 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr4_-_102268628 | 0.06 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_193155729 | 0.06 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr19_+_46001697 | 0.06 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.4 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.2 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) epinephrine secretion(GO:0048242) |
| 0.1 | 0.2 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.0 | GO:0072185 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.0 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0060264 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:0061150 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0072034 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0098519 | phosphoserine phosphatase activity(GO:0004647) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |