Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
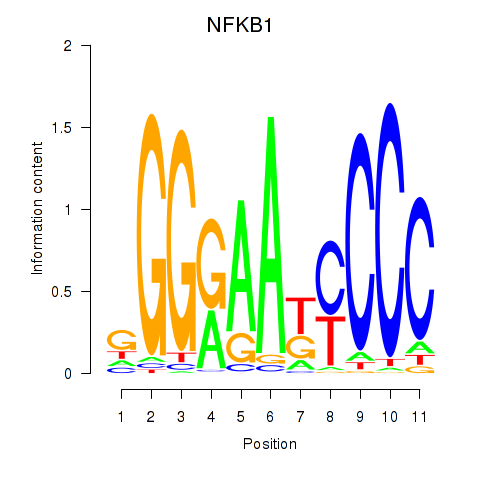
Results for NFKB1
Z-value: 0.72
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.7 | nuclear factor kappa B subunit 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg19_v2_chr4_+_103422499_103422632 | 0.85 | 1.5e-01 | Click! |
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39735646 | 0.62 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr19_+_39759154 | 0.53 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr6_+_138188351 | 0.50 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_+_6845578 | 0.43 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr4_-_74904398 | 0.41 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr2_-_61108449 | 0.40 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr4_+_103422499 | 0.36 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr4_+_74735102 | 0.35 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr6_-_31550192 | 0.34 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr12_-_49259643 | 0.30 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr9_+_82188077 | 0.30 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_+_6845497 | 0.28 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr17_-_77179487 | 0.27 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr2_+_228678550 | 0.26 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr17_-_61776522 | 0.25 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr7_+_143013198 | 0.24 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr16_-_66959429 | 0.24 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr11_+_94706804 | 0.23 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr15_-_89438742 | 0.23 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr10_+_12391685 | 0.22 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr6_+_138188551 | 0.20 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_+_39574945 | 0.19 |
ENST00000331256.5
|
PAPL
|
Iron/zinc purple acid phosphatase-like protein |
| chr20_+_6748311 | 0.19 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr6_+_144471643 | 0.19 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr2_-_211036051 | 0.19 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr14_-_51297360 | 0.18 |
ENST00000496749.1
|
NIN
|
ninein (GSK3B interacting protein) |
| chr1_+_56880606 | 0.18 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr12_-_12849073 | 0.18 |
ENST00000332427.2
ENST00000540796.1 |
GPR19
|
G protein-coupled receptor 19 |
| chr4_-_5891918 | 0.18 |
ENST00000512574.1
|
CRMP1
|
collapsin response mediator protein 1 |
| chr2_+_176957619 | 0.17 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr12_+_52445191 | 0.17 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_+_29823427 | 0.17 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr2_+_29001711 | 0.17 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_38100539 | 0.17 |
ENST00000401069.1
|
RSPO1
|
R-spondin 1 |
| chr12_+_7053228 | 0.17 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr14_-_35873856 | 0.17 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr2_+_112895939 | 0.17 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr5_+_170846640 | 0.17 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr1_-_8000872 | 0.16 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr6_-_29527702 | 0.16 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr20_-_44937124 | 0.16 |
ENST00000537909.1
|
CDH22
|
cadherin 22, type 2 |
| chr14_+_96343100 | 0.16 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr4_-_74964904 | 0.15 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr17_+_46189311 | 0.15 |
ENST00000582481.1
|
SNX11
|
sorting nexin 11 |
| chr20_+_44746939 | 0.15 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr9_-_140115775 | 0.15 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr2_-_42721110 | 0.15 |
ENST00000394973.4
ENST00000306078.1 |
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr8_-_103668114 | 0.15 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr20_-_62103862 | 0.15 |
ENST00000344462.4
ENST00000357249.2 ENST00000359125.2 ENST00000360480.3 ENST00000370224.1 ENST00000344425.5 ENST00000354587.3 ENST00000359689.1 |
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2 |
| chr6_+_138188378 | 0.15 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr6_-_35464727 | 0.14 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr1_-_161168834 | 0.14 |
ENST00000367995.3
ENST00000367996.5 |
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
| chr7_-_56101826 | 0.14 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr3_-_111852061 | 0.14 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr4_-_147442982 | 0.14 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr14_+_96342729 | 0.14 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr19_+_2476116 | 0.14 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr20_-_42816206 | 0.14 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr1_+_100818156 | 0.13 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr11_+_94706973 | 0.13 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr1_+_26872324 | 0.13 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr12_-_62997214 | 0.13 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr12_-_58131931 | 0.13 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr2_-_157189180 | 0.12 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_+_99609996 | 0.12 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr17_+_21191341 | 0.12 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr14_+_32798547 | 0.12 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr13_+_108921977 | 0.12 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr2_-_193059250 | 0.12 |
ENST00000409056.3
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr4_-_74864386 | 0.12 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr12_+_7053172 | 0.12 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr10_+_104154229 | 0.12 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr6_-_24720226 | 0.12 |
ENST00000378102.3
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr17_+_4854375 | 0.12 |
ENST00000521811.1
ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3
|
enolase 3 (beta, muscle) |
| chr8_+_26149007 | 0.11 |
ENST00000380737.3
ENST00000524169.1 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr15_+_33010175 | 0.11 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr19_+_45504688 | 0.11 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr1_-_155658085 | 0.11 |
ENST00000311573.5
ENST00000438245.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr19_+_10527449 | 0.11 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr7_+_120969045 | 0.11 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr4_-_147443043 | 0.11 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr17_-_56065540 | 0.10 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr16_-_28222797 | 0.10 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr10_-_6104253 | 0.10 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr6_-_44233361 | 0.10 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr6_-_35464817 | 0.10 |
ENST00000338863.7
|
TEAD3
|
TEA domain family member 3 |
| chr16_-_77468945 | 0.10 |
ENST00000282849.5
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr2_+_64681641 | 0.10 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr12_-_102455846 | 0.10 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr7_-_93520191 | 0.10 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr12_+_100661156 | 0.09 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr11_+_102188272 | 0.09 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr20_+_18488528 | 0.09 |
ENST00000377465.1
|
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr11_+_102188224 | 0.09 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chrX_-_38080077 | 0.09 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr20_+_18488137 | 0.09 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr19_-_51920952 | 0.09 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr4_-_185395672 | 0.09 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr15_+_89346699 | 0.09 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr5_-_55290773 | 0.09 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr19_+_17638041 | 0.09 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr19_-_39390350 | 0.09 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr3_+_159557637 | 0.09 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr4_-_5890145 | 0.09 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr19_-_39390440 | 0.09 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr17_-_75878647 | 0.09 |
ENST00000374983.2
|
FLJ45079
|
FLJ45079 |
| chr6_-_31138439 | 0.09 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr6_-_30523865 | 0.09 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr16_+_67465016 | 0.09 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr11_-_77185094 | 0.09 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr19_+_42364460 | 0.08 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr12_-_57081940 | 0.08 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr3_+_10857885 | 0.08 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr1_-_236030216 | 0.08 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr1_-_186649543 | 0.08 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_93520259 | 0.08 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr2_+_163175394 | 0.08 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr1_-_204436344 | 0.08 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr1_-_36615065 | 0.08 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr16_-_28223166 | 0.08 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr15_-_66545995 | 0.08 |
ENST00000395614.1
ENST00000288745.3 ENST00000422354.1 ENST00000395625.2 ENST00000360698.4 ENST00000409699.2 |
MEGF11
|
multiple EGF-like-domains 11 |
| chr19_+_55795493 | 0.08 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chrX_-_37706815 | 0.08 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr1_-_32403903 | 0.08 |
ENST00000344035.6
ENST00000356536.3 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr9_-_115480303 | 0.08 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr1_+_159796534 | 0.08 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr20_+_44746885 | 0.08 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr11_-_46142615 | 0.08 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chrX_-_73072534 | 0.08 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr3_-_111852128 | 0.08 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chr14_-_61190754 | 0.07 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr10_-_97050777 | 0.07 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr6_+_14117872 | 0.07 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr3_-_4793274 | 0.07 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr10_+_13142075 | 0.07 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr5_-_150460914 | 0.07 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_+_103422471 | 0.07 |
ENST00000226574.4
ENST00000394820.4 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr10_-_13390021 | 0.07 |
ENST00000537130.1
|
SEPHS1
|
selenophosphate synthetase 1 |
| chr15_-_66084621 | 0.07 |
ENST00000564674.1
|
DENND4A
|
DENN/MADD domain containing 4A |
| chr19_-_51920835 | 0.07 |
ENST00000442846.3
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr14_+_24521202 | 0.07 |
ENST00000334420.7
ENST00000342740.5 |
LRRC16B
|
leucine rich repeat containing 16B |
| chr17_-_61819229 | 0.07 |
ENST00000447001.3
ENST00000392950.4 |
STRADA
|
STE20-related kinase adaptor alpha |
| chr9_-_127952187 | 0.07 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr6_-_30080863 | 0.07 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr10_+_13141585 | 0.07 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr19_+_17638059 | 0.06 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr17_-_61819121 | 0.06 |
ENST00000245865.5
ENST00000375840.4 ENST00000582137.1 ENST00000579549.1 ENST00000582030.1 ENST00000584110.1 ENST00000580288.1 ENST00000336174.6 ENST00000579340.1 ENST00000580338.1 |
STRADA
|
STE20-related kinase adaptor alpha |
| chr1_-_36615051 | 0.06 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr3_+_52279902 | 0.06 |
ENST00000457454.1
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr16_+_28834531 | 0.06 |
ENST00000570200.1
|
ATXN2L
|
ataxin 2-like |
| chr19_-_51920873 | 0.06 |
ENST00000441969.3
ENST00000525998.1 ENST00000436984.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr1_+_155657737 | 0.06 |
ENST00000471642.2
ENST00000471214.1 |
DAP3
|
death associated protein 3 |
| chr12_-_54813229 | 0.06 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr17_-_61777090 | 0.06 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr6_-_31612808 | 0.06 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr4_+_142557771 | 0.06 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr14_+_75536335 | 0.06 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr6_-_74230741 | 0.06 |
ENST00000316292.9
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr20_+_1115821 | 0.06 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr9_+_33817461 | 0.06 |
ENST00000263228.3
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr2_-_136743436 | 0.06 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr3_+_63638372 | 0.06 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chrX_+_117861535 | 0.06 |
ENST00000371666.3
ENST00000371642.1 |
IL13RA1
|
interleukin 13 receptor, alpha 1 |
| chrX_+_40440146 | 0.06 |
ENST00000535539.1
ENST00000378438.4 ENST00000436783.1 ENST00000544975.1 ENST00000535777.1 ENST00000447485.1 ENST00000423649.1 |
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr6_+_163835669 | 0.06 |
ENST00000453779.2
ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI
|
QKI, KH domain containing, RNA binding |
| chr1_-_113249678 | 0.06 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr5_-_150521192 | 0.06 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr20_-_62462566 | 0.06 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr4_-_120549163 | 0.06 |
ENST00000394439.1
ENST00000420633.1 |
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr4_-_76598326 | 0.06 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_-_150039249 | 0.05 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr5_-_127418755 | 0.05 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr2_+_202047843 | 0.05 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr14_-_24615805 | 0.05 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr10_+_89124746 | 0.05 |
ENST00000465545.1
|
NUTM2D
|
NUT family member 2D |
| chrX_-_15872914 | 0.05 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_+_75668916 | 0.05 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr1_+_154301264 | 0.05 |
ENST00000341822.2
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr15_+_22892663 | 0.05 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr1_+_89990431 | 0.05 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr13_+_97874574 | 0.05 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr1_+_205012293 | 0.05 |
ENST00000331830.4
|
CNTN2
|
contactin 2 (axonal) |
| chr9_+_82187487 | 0.05 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr3_-_194072019 | 0.05 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr1_+_62417957 | 0.05 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr1_+_38259459 | 0.05 |
ENST00000373045.6
|
MANEAL
|
mannosidase, endo-alpha-like |
| chr1_-_54304212 | 0.05 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr1_+_201979743 | 0.05 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr6_-_17706618 | 0.05 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr9_-_136344197 | 0.05 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr12_-_48499591 | 0.05 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr10_+_89419370 | 0.05 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr1_+_27114418 | 0.05 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr17_+_7590734 | 0.05 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr22_+_38349724 | 0.05 |
ENST00000470701.1
|
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr10_-_51623203 | 0.05 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr19_+_4474846 | 0.05 |
ENST00000589486.1
ENST00000592691.1 |
HDGFRP2
|
Hepatoma-derived growth factor-related protein 2 |
| chr6_+_18155632 | 0.05 |
ENST00000297792.5
|
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr1_+_10093188 | 0.04 |
ENST00000377153.1
|
UBE4B
|
ubiquitination factor E4B |
| chr1_+_110453462 | 0.04 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.4 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.1 | 0.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.3 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.2 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.1 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.1 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.0 | 0.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:1990619 | growth involved in heart morphogenesis(GO:0003241) positive regulation of chondrocyte proliferation(GO:1902732) histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0097476 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.0 | GO:1904437 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.0 | GO:0060168 | regulation of axon diameter(GO:0031133) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.0 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |