Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
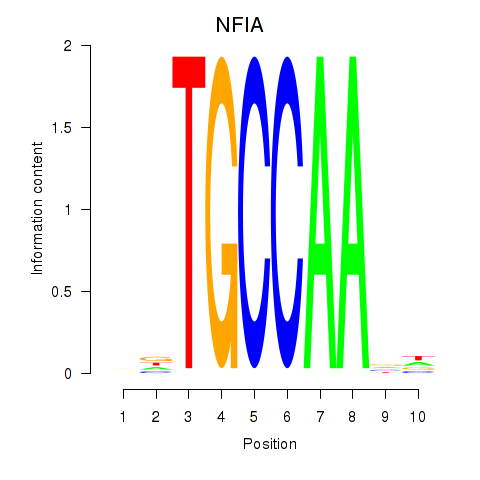
Results for NFIA
Z-value: 1.83
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.11 | nuclear factor I A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg19_v2_chr1_+_61548225_61548299 | -0.89 | 1.1e-01 | Click! |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_95975672 | 1.06 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr4_-_157892055 | 0.86 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr14_+_77582905 | 0.86 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr5_-_34043310 | 0.80 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr1_+_112016414 | 0.79 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr14_-_98444386 | 0.76 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chrX_-_71497148 | 0.74 |
ENST00000316084.6
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr1_-_114429997 | 0.66 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr12_-_13248598 | 0.66 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr9_+_130911770 | 0.65 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr7_+_79764104 | 0.58 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr7_-_151330218 | 0.57 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_+_135340859 | 0.57 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr18_+_61144160 | 0.57 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr7_-_17598506 | 0.56 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr11_-_122931881 | 0.55 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr17_+_67957878 | 0.53 |
ENST00000420427.1
|
AC004562.1
|
AC004562.1 |
| chr8_+_66619277 | 0.53 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr11_+_111749650 | 0.53 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr5_-_156362666 | 0.52 |
ENST00000406964.1
|
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr12_+_48225126 | 0.52 |
ENST00000550909.1
ENST00000550720.1 ENST00000548564.1 |
RP5-1057I20.2
|
RP5-1057I20.2 |
| chr11_+_77774897 | 0.52 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr19_+_42254885 | 0.52 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr2_+_3705785 | 0.51 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr3_-_105588231 | 0.50 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_+_33701684 | 0.49 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr13_-_100624012 | 0.48 |
ENST00000267294.4
|
ZIC5
|
Zic family member 5 |
| chrX_-_119693745 | 0.46 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr3_-_52488048 | 0.46 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr6_-_10115007 | 0.46 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr5_+_74807581 | 0.45 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chrX_-_69479654 | 0.45 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr8_+_124194875 | 0.45 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr21_+_44394742 | 0.45 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr5_-_55008136 | 0.45 |
ENST00000503891.1
ENST00000507109.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr5_-_43313574 | 0.44 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr12_+_110718428 | 0.44 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr2_+_136289030 | 0.43 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr12_+_100897130 | 0.43 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_180018540 | 0.43 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr5_-_168006324 | 0.43 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chrX_-_71497077 | 0.41 |
ENST00000373626.3
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr13_-_36705425 | 0.41 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr8_-_71519889 | 0.40 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr12_+_56435637 | 0.40 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr1_-_204119009 | 0.39 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2 |
| chr7_-_27135591 | 0.38 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr4_+_146402346 | 0.37 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr2_-_180871780 | 0.37 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr11_-_93271058 | 0.37 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr18_+_20494078 | 0.37 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr5_+_112074029 | 0.37 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr6_+_13925098 | 0.37 |
ENST00000488300.1
ENST00000544682.1 ENST00000420478.2 |
RNF182
|
ring finger protein 182 |
| chr14_-_38036271 | 0.37 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr7_+_129932974 | 0.36 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr16_-_21442874 | 0.36 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr14_-_69350920 | 0.36 |
ENST00000553290.1
|
ACTN1
|
actinin, alpha 1 |
| chr12_+_93965609 | 0.36 |
ENST00000549887.1
ENST00000551556.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr12_+_110562135 | 0.36 |
ENST00000361948.4
ENST00000552912.1 ENST00000242591.5 ENST00000546374.1 |
IFT81
|
intraflagellar transport 81 homolog (Chlamydomonas) |
| chr9_+_123906331 | 0.35 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr3_+_52350335 | 0.35 |
ENST00000420323.2
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr1_+_170501270 | 0.35 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr2_-_69180083 | 0.35 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr5_+_112073544 | 0.34 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr4_-_111558135 | 0.34 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr17_-_37764128 | 0.34 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr14_-_64194745 | 0.34 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr3_+_32737027 | 0.34 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr6_-_86353510 | 0.34 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr5_-_43313269 | 0.34 |
ENST00000511774.1
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr8_+_97597148 | 0.34 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr8_+_104384616 | 0.34 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr6_-_2971494 | 0.34 |
ENST00000380539.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr17_+_65375082 | 0.34 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr8_+_32579271 | 0.33 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr13_+_78109804 | 0.33 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr17_+_1733276 | 0.33 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr17_-_40169161 | 0.32 |
ENST00000589586.2
ENST00000426588.3 ENST00000589576.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr11_-_113644491 | 0.32 |
ENST00000200135.3
|
ZW10
|
zw10 kinetochore protein |
| chr17_+_54671047 | 0.32 |
ENST00000332822.4
|
NOG
|
noggin |
| chr13_+_78109884 | 0.32 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr11_+_33037652 | 0.31 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr14_-_50319758 | 0.31 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr17_-_42188598 | 0.31 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr22_-_23922410 | 0.31 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr1_+_153746683 | 0.31 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr5_-_55008101 | 0.31 |
ENST00000506624.1
ENST00000513275.1 ENST00000513993.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr2_-_61244550 | 0.30 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr15_+_41549105 | 0.30 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr2_-_136288740 | 0.30 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr17_+_1733251 | 0.30 |
ENST00000570451.1
|
RPA1
|
replication protein A1, 70kDa |
| chr12_-_21910775 | 0.30 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr18_+_61143994 | 0.30 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr17_-_39459103 | 0.30 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr1_-_154164534 | 0.30 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr8_+_124194752 | 0.30 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr8_+_102064237 | 0.29 |
ENST00000514926.1
|
RP11-302J23.1
|
RP11-302J23.1 |
| chr2_+_33701707 | 0.29 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr3_+_159570722 | 0.29 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr16_-_15474904 | 0.29 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr3_-_131736593 | 0.29 |
ENST00000514999.1
|
CPNE4
|
copine IV |
| chr2_-_211036051 | 0.29 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr15_+_36994210 | 0.29 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr11_+_34654011 | 0.29 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr3_-_93747425 | 0.29 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr22_+_38864041 | 0.29 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr4_+_76439095 | 0.28 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr1_+_68150744 | 0.28 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr3_+_152017360 | 0.28 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr3_+_142342240 | 0.28 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr3_+_150264458 | 0.28 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr2_+_62132781 | 0.28 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr5_+_89770696 | 0.28 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chrX_-_19817869 | 0.27 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr5_+_122847908 | 0.27 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr17_-_15168624 | 0.27 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr12_-_53207842 | 0.27 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr12_-_95510743 | 0.27 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr4_-_113437328 | 0.27 |
ENST00000313341.3
|
NEUROG2
|
neurogenin 2 |
| chr13_+_23755054 | 0.27 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr9_+_137967366 | 0.27 |
ENST00000252854.4
|
OLFM1
|
olfactomedin 1 |
| chr18_-_12884259 | 0.27 |
ENST00000353319.4
ENST00000327283.3 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr1_-_235098935 | 0.27 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr3_-_52002194 | 0.27 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr9_+_17134980 | 0.27 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr17_-_47786375 | 0.27 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr9_-_129885010 | 0.27 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr1_+_15986364 | 0.27 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr6_+_43968306 | 0.26 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr5_-_142783694 | 0.26 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr2_+_54684327 | 0.26 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr6_+_126221034 | 0.26 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chrX_+_135252050 | 0.25 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr19_-_40724246 | 0.25 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr3_-_25824872 | 0.25 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr10_-_13390021 | 0.25 |
ENST00000537130.1
|
SEPHS1
|
selenophosphate synthetase 1 |
| chr2_-_114300213 | 0.25 |
ENST00000446595.1
ENST00000416105.1 ENST00000450636.1 ENST00000416758.1 |
RP11-395L14.4
|
RP11-395L14.4 |
| chr13_-_46425865 | 0.25 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr14_+_55494323 | 0.25 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr11_+_62556596 | 0.25 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr8_-_16859690 | 0.24 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr11_-_122930121 | 0.24 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr10_-_14050522 | 0.24 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr9_+_17135016 | 0.24 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr11_+_34643600 | 0.24 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr11_-_64013288 | 0.24 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_236046872 | 0.24 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr3_+_69788576 | 0.24 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_13461790 | 0.24 |
ENST00000530907.1
|
BTBD10
|
BTB (POZ) domain containing 10 |
| chr4_+_146402997 | 0.24 |
ENST00000512019.1
|
SMAD1
|
SMAD family member 1 |
| chr5_-_74807418 | 0.24 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr10_-_5227096 | 0.24 |
ENST00000488756.1
ENST00000334314.3 |
AKR1CL1
|
aldo-keto reductase family 1, member C-like 1 |
| chr20_-_14318248 | 0.23 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chrX_-_135849484 | 0.23 |
ENST00000370620.1
ENST00000535227.1 |
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr17_+_79031415 | 0.23 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr10_-_61122220 | 0.23 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr5_+_122847781 | 0.23 |
ENST00000395412.1
ENST00000395411.1 ENST00000345990.4 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr7_+_17338239 | 0.23 |
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor |
| chr19_-_1812193 | 0.23 |
ENST00000525591.1
|
ATP8B3
|
ATPase, aminophospholipid transporter, class I, type 8B, member 3 |
| chr17_-_67224812 | 0.23 |
ENST00000423818.2
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10 |
| chr1_-_114430169 | 0.23 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chrX_+_122993657 | 0.23 |
ENST00000434753.3
ENST00000430625.1 |
XIAP
|
X-linked inhibitor of apoptosis |
| chr3_+_109128837 | 0.23 |
ENST00000497996.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr8_+_121457642 | 0.23 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr4_+_146402925 | 0.23 |
ENST00000302085.4
|
SMAD1
|
SMAD family member 1 |
| chr6_+_13925318 | 0.23 |
ENST00000423553.2
ENST00000537388.1 |
RNF182
|
ring finger protein 182 |
| chr12_-_82752565 | 0.22 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr2_-_219925189 | 0.22 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chrX_-_106146547 | 0.22 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr12_-_99038732 | 0.22 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr14_+_70234642 | 0.22 |
ENST00000555349.1
ENST00000554021.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr14_+_55493920 | 0.22 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr22_+_24129138 | 0.22 |
ENST00000417137.1
ENST00000344921.6 ENST00000263121.7 ENST00000407422.3 ENST00000407082.3 |
SMARCB1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
| chr10_-_27149851 | 0.22 |
ENST00000376142.2
ENST00000359188.4 ENST00000376139.2 ENST00000376160.1 |
ABI1
|
abl-interactor 1 |
| chr19_-_12997995 | 0.22 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr16_+_22518495 | 0.22 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr11_+_193065 | 0.22 |
ENST00000342878.2
|
SCGB1C1
|
secretoglobin, family 1C, member 1 |
| chr6_+_42989344 | 0.22 |
ENST00000244496.5
|
RRP36
|
ribosomal RNA processing 36 homolog (S. cerevisiae) |
| chr6_-_32784687 | 0.22 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr17_+_27369918 | 0.22 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr5_-_43515231 | 0.22 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr2_+_192543694 | 0.21 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr1_-_160253998 | 0.21 |
ENST00000392220.2
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr7_+_107332334 | 0.21 |
ENST00000541474.1
ENST00000544569.1 |
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr3_-_37216055 | 0.21 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_43864387 | 0.21 |
ENST00000282406.4
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr12_-_82752159 | 0.21 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr1_+_66797687 | 0.21 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr11_-_61348292 | 0.21 |
ENST00000539008.1
ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7
|
synaptotagmin VII |
| chr5_-_55777586 | 0.21 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr16_+_82090028 | 0.21 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr11_-_125550726 | 0.21 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chrX_-_85302531 | 0.21 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr11_-_85430204 | 0.21 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr15_+_38544476 | 0.20 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr3_+_186501979 | 0.20 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr3_+_152017924 | 0.20 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr11_-_85430088 | 0.20 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_+_46379254 | 0.20 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr7_+_35756092 | 0.20 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.2 | 0.8 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.2 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.7 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.5 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 1.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.4 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.5 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.6 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.4 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.3 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.3 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.3 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.7 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.2 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.5 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.3 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.6 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle cell proliferation(GO:0014858) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.2 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.1 | 0.2 | GO:1990927 | short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 1.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.2 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.8 | GO:0009642 | response to light intensity(GO:0009642) response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:1902510 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.4 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 1.1 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0014740 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0060294 | inner dynein arm assembly(GO:0036159) cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.2 | GO:0097324 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.4 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 2.0 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.6 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.3 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.4 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.4 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |