Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
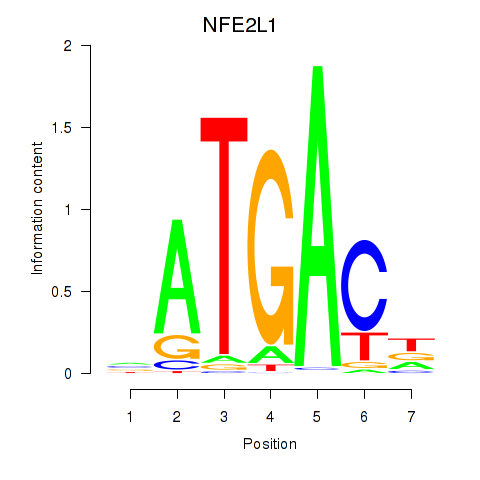
Results for NFE2L1
Z-value: 0.70
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | nuclear factor, erythroid 2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46132037_46132080 | 0.95 | 4.8e-02 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_34665983 | 0.52 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr10_-_75168071 | 0.51 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr14_+_24779340 | 0.49 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr11_+_63742050 | 0.42 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr9_-_128246769 | 0.41 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr16_+_3068393 | 0.41 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr15_+_80215113 | 0.38 |
ENST00000560255.1
|
C15orf37
|
chromosome 15 open reading frame 37 |
| chr16_+_21623958 | 0.38 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr15_-_54267147 | 0.36 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr19_+_42773371 | 0.33 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr19_+_55996316 | 0.31 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr11_+_118754475 | 0.31 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr8_+_22853345 | 0.30 |
ENST00000522948.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr6_+_27114861 | 0.30 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr4_-_89442940 | 0.28 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr11_+_2397418 | 0.28 |
ENST00000530648.1
|
CD81
|
CD81 molecule |
| chr8_+_94752349 | 0.28 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr19_-_36545649 | 0.27 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr14_+_24779376 | 0.27 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chrX_+_47077680 | 0.27 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr22_+_30805086 | 0.26 |
ENST00000439838.1
ENST00000439023.3 |
RP4-539M6.19
|
Uncharacterized protein |
| chr6_+_30029008 | 0.25 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr19_-_42721819 | 0.25 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr11_+_65337901 | 0.24 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr9_-_130477912 | 0.24 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr2_-_98280383 | 0.23 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr3_-_172019686 | 0.23 |
ENST00000596321.1
|
AC092964.2
|
Uncharacterized protein |
| chr6_-_154751629 | 0.23 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr11_-_64052111 | 0.23 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr19_+_18699535 | 0.23 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr12_+_51633061 | 0.22 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr17_+_73606766 | 0.22 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr3_-_47950745 | 0.22 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr17_+_38375528 | 0.22 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr14_-_69262947 | 0.22 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_+_9361606 | 0.22 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr5_-_180242534 | 0.22 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr14_+_23790655 | 0.21 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr5_+_149877440 | 0.21 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_+_143381594 | 0.21 |
ENST00000367601.4
|
AIG1
|
androgen-induced 1 |
| chr9_-_140082983 | 0.21 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr1_+_81106951 | 0.20 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr20_+_43992094 | 0.20 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr9_+_140083099 | 0.20 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr20_-_36794902 | 0.20 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr3_-_114790179 | 0.20 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_74685413 | 0.20 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr16_-_1470709 | 0.20 |
ENST00000563974.1
ENST00000442039.2 |
C16orf91
|
chromosome 16 open reading frame 91 |
| chr1_+_100435986 | 0.20 |
ENST00000532693.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr14_+_102276192 | 0.20 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chrX_+_47078069 | 0.19 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr21_-_33975547 | 0.19 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr11_-_62380199 | 0.19 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr5_-_180242576 | 0.18 |
ENST00000514438.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr17_-_7145106 | 0.18 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr1_+_45274154 | 0.18 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr22_-_38349552 | 0.18 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr1_+_159750776 | 0.18 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr6_-_32145861 | 0.18 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr6_+_32146268 | 0.18 |
ENST00000427134.2
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr15_+_89182156 | 0.17 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr3_-_47934234 | 0.17 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr19_-_44100275 | 0.17 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr8_-_144623595 | 0.17 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chr6_-_31940065 | 0.17 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr5_+_139055055 | 0.17 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr1_+_159750720 | 0.17 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr19_+_44100632 | 0.17 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr11_+_64052266 | 0.17 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr11_+_64052294 | 0.17 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr14_+_38033252 | 0.17 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr19_-_50370799 | 0.16 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr5_+_175223313 | 0.16 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr20_+_327413 | 0.16 |
ENST00000609179.1
|
NRSN2
|
neurensin 2 |
| chr4_-_40632757 | 0.16 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr9_+_131799213 | 0.16 |
ENST00000358369.4
ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B
|
family with sequence similarity 73, member B |
| chr16_+_29819446 | 0.16 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr22_-_30968813 | 0.16 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr14_-_75083313 | 0.16 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr5_+_139055021 | 0.16 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr11_+_64052454 | 0.16 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr6_+_33378738 | 0.15 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr16_-_75301886 | 0.15 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr7_+_127527965 | 0.14 |
ENST00000486037.1
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr22_-_30968839 | 0.14 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr8_+_107738343 | 0.14 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr6_+_89790490 | 0.14 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr16_+_57662596 | 0.14 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_+_18699599 | 0.14 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr19_-_12807422 | 0.13 |
ENST00000380339.3
ENST00000544494.1 ENST00000393261.3 |
FBXW9
|
F-box and WD repeat domain containing 9 |
| chr20_-_44516256 | 0.13 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr9_+_134065506 | 0.13 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr14_+_24769043 | 0.13 |
ENST00000267425.3
ENST00000396802.3 |
NOP9
|
NOP9 nucleolar protein |
| chr11_+_63137251 | 0.13 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chrX_-_148713440 | 0.13 |
ENST00000536359.1
ENST00000316916.8 |
TMEM185A
|
transmembrane protein 185A |
| chr18_+_72166564 | 0.13 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr4_-_40632881 | 0.13 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_-_71223839 | 0.12 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr19_-_47616992 | 0.12 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr10_-_51958906 | 0.12 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr1_+_63788730 | 0.12 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr11_-_62477103 | 0.12 |
ENST00000532818.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr4_+_8442496 | 0.12 |
ENST00000389737.4
ENST00000504134.1 |
TRMT44
|
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr2_+_27301435 | 0.12 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr17_-_73663245 | 0.12 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chrX_-_84363974 | 0.12 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr17_+_42264395 | 0.11 |
ENST00000587989.1
ENST00000590235.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr12_+_49658855 | 0.11 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr5_+_140248518 | 0.11 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr12_+_57998400 | 0.11 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr5_-_162887054 | 0.11 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr11_+_64950801 | 0.11 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr17_+_19186292 | 0.11 |
ENST00000395626.1
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr11_-_125366089 | 0.11 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_+_111580508 | 0.11 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr4_+_4861385 | 0.11 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr7_+_6713376 | 0.11 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr3_+_112929850 | 0.11 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr11_+_35222629 | 0.11 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr18_-_2982869 | 0.11 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr11_+_117063295 | 0.11 |
ENST00000525478.1
ENST00000532062.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr1_-_151319654 | 0.11 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr7_-_11871815 | 0.11 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr17_+_42264556 | 0.11 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr3_+_47422485 | 0.11 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr20_-_4229721 | 0.11 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr1_+_28586006 | 0.11 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr15_+_101389945 | 0.10 |
ENST00000561231.1
ENST00000559331.1 ENST00000558254.1 |
RP11-66B24.2
|
RP11-66B24.2 |
| chr19_+_44100727 | 0.10 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr1_-_12679171 | 0.10 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr1_-_24126051 | 0.10 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr17_+_7155819 | 0.10 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr1_+_67632083 | 0.10 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr4_-_40632844 | 0.10 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr7_+_871559 | 0.10 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr10_-_49860525 | 0.10 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr11_-_66103932 | 0.10 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_+_65154070 | 0.10 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr17_-_73663168 | 0.10 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr3_-_118753792 | 0.10 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr12_+_6644443 | 0.10 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr19_-_45681482 | 0.10 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr2_+_74685527 | 0.10 |
ENST00000393972.3
ENST00000409737.1 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr7_+_150811705 | 0.10 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr2_+_159651821 | 0.10 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr3_-_120365866 | 0.09 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr22_-_39928823 | 0.09 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr2_-_228244013 | 0.09 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr9_+_35538616 | 0.09 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr17_-_27044760 | 0.09 |
ENST00000395243.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr1_+_180875711 | 0.09 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr17_+_76210367 | 0.09 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr22_+_42229100 | 0.09 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr6_-_31939734 | 0.09 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr1_-_183538319 | 0.09 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr21_-_35773370 | 0.09 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr9_+_131901661 | 0.09 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr22_+_38142235 | 0.09 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr2_-_25451065 | 0.09 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr9_+_139847347 | 0.09 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr6_+_131958436 | 0.09 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr11_-_66103867 | 0.09 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr12_+_22778291 | 0.09 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr11_+_128563652 | 0.08 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr12_+_72080253 | 0.08 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr4_-_103749179 | 0.08 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_31107127 | 0.08 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr7_-_34978980 | 0.08 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr10_+_102759045 | 0.08 |
ENST00000370220.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr17_+_26662679 | 0.08 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr2_-_230787879 | 0.08 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr17_+_6916527 | 0.08 |
ENST00000552321.1
|
RNASEK
|
ribonuclease, RNase K |
| chr9_-_116840728 | 0.08 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr17_+_42264322 | 0.08 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_+_74701062 | 0.08 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr4_-_159080806 | 0.08 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr15_+_28624878 | 0.08 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr9_+_134065519 | 0.08 |
ENST00000531600.1
|
NUP214
|
nucleoporin 214kDa |
| chr6_-_33297013 | 0.08 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr6_-_43027105 | 0.08 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr16_+_57662419 | 0.08 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chrX_+_49126294 | 0.07 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr2_-_219524193 | 0.07 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
| chr1_-_24126023 | 0.07 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr2_+_54198210 | 0.07 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr6_+_26204825 | 0.07 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr2_+_113033164 | 0.07 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr1_+_46806452 | 0.07 |
ENST00000536062.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr16_+_67906919 | 0.07 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr19_-_46142637 | 0.07 |
ENST00000590043.1
ENST00000589876.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_+_41856816 | 0.07 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr11_+_126139005 | 0.07 |
ENST00000263578.5
ENST00000442061.2 ENST00000532125.1 |
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1 |
| chrX_+_15767971 | 0.07 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr17_-_56591978 | 0.07 |
ENST00000583656.1
|
MTMR4
|
myotubularin related protein 4 |
| chr1_+_32645269 | 0.07 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr14_-_77542485 | 0.07 |
ENST00000556781.1
ENST00000557526.1 ENST00000555512.1 |
RP11-7F17.3
|
RP11-7F17.3 |
| chr8_-_95220775 | 0.07 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr19_-_14606900 | 0.07 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr10_-_69991865 | 0.07 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr6_+_168418553 | 0.07 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.2 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0044533 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.3 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.6 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.3 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |