Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
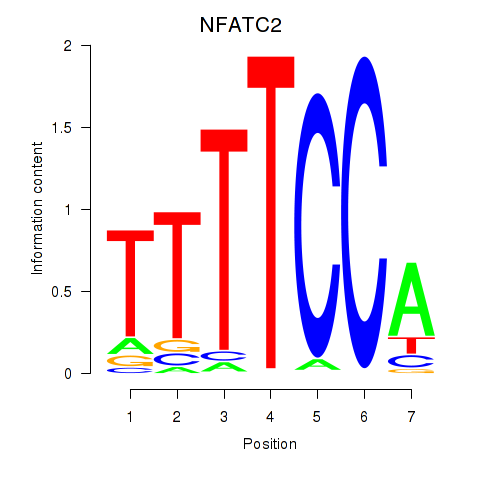
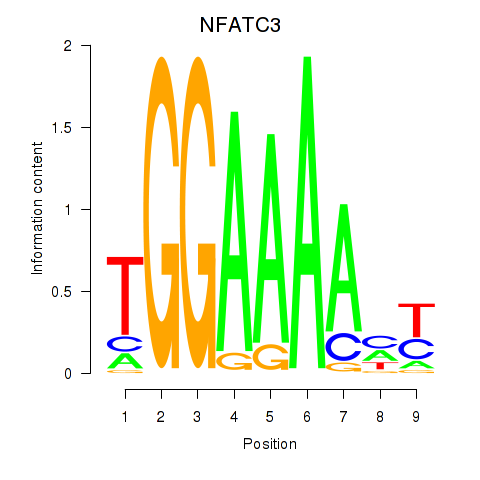
Results for NFATC2_NFATC3
Z-value: 0.54
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.15 | nuclear factor of activated T cells 2 |
|
NFATC3
|
ENSG00000072736.14 | nuclear factor of activated T cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC3 | hg19_v2_chr16_+_68119247_68119293 | -0.79 | 2.1e-01 | Click! |
| NFATC2 | hg19_v2_chr20_-_50179368_50179392 | -0.62 | 3.8e-01 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_35107346 | 0.45 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr2_+_162272605 | 0.28 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr17_+_18647326 | 0.26 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr17_+_42925270 | 0.25 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr6_+_26183958 | 0.23 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chrX_-_15683147 | 0.22 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr17_-_15469590 | 0.21 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr5_-_78808617 | 0.21 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chrX_-_139866723 | 0.21 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr4_+_39408470 | 0.20 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr19_-_39735646 | 0.19 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr6_-_26235206 | 0.19 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr8_+_66619277 | 0.18 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr14_+_32546274 | 0.17 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr3_+_69812877 | 0.16 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr16_-_28192360 | 0.16 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr5_+_131993856 | 0.16 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr11_+_128563652 | 0.15 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr19_+_18726786 | 0.15 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr11_-_64527425 | 0.14 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr3_+_140981456 | 0.14 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr8_-_23712312 | 0.14 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr17_-_46690839 | 0.14 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr8_+_86121448 | 0.14 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr6_+_138188351 | 0.14 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr9_-_21077939 | 0.13 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr11_+_9482551 | 0.13 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr6_+_138188378 | 0.13 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_+_90864504 | 0.13 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_+_38022572 | 0.12 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr4_-_74847800 | 0.12 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr10_-_50323543 | 0.12 |
ENST00000332853.4
ENST00000298454.3 |
VSTM4
|
V-set and transmembrane domain containing 4 |
| chr12_+_49961990 | 0.11 |
ENST00000551063.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr1_-_91487013 | 0.11 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr7_+_127881325 | 0.11 |
ENST00000308868.4
|
LEP
|
leptin |
| chr6_+_26156551 | 0.11 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr11_-_69867159 | 0.11 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr2_+_232316906 | 0.11 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr13_+_113030658 | 0.11 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr12_+_79357815 | 0.11 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr19_+_7599792 | 0.11 |
ENST00000600942.1
ENST00000593924.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr14_+_77582905 | 0.10 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr8_+_52730143 | 0.10 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr3_-_167191814 | 0.10 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr12_-_49582978 | 0.10 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_-_134495992 | 0.10 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr13_-_30160925 | 0.10 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_+_105036909 | 0.10 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr15_+_84841242 | 0.10 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr6_+_155538093 | 0.10 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr17_-_8263538 | 0.10 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr1_+_209941942 | 0.10 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr6_+_76599809 | 0.10 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr11_+_22696314 | 0.09 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr12_-_49582593 | 0.09 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr12_-_50616122 | 0.09 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr17_+_7621045 | 0.09 |
ENST00000570791.1
|
DNAH2
|
dynein, axonemal, heavy chain 2 |
| chr19_+_7599597 | 0.09 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr14_-_52535712 | 0.09 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr13_+_37247934 | 0.09 |
ENST00000315190.3
|
SERTM1
|
serine-rich and transmembrane domain containing 1 |
| chr1_+_86046433 | 0.09 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr3_+_152017360 | 0.09 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr11_+_9482512 | 0.09 |
ENST00000396602.2
ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143
|
zinc finger protein 143 |
| chr1_-_46152174 | 0.09 |
ENST00000290795.3
ENST00000355105.3 |
GPBP1L1
|
GC-rich promoter binding protein 1-like 1 |
| chr1_+_44401479 | 0.09 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr7_+_102553430 | 0.09 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr17_-_61776522 | 0.09 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr11_+_111749650 | 0.09 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr5_+_74011328 | 0.09 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr12_-_100656134 | 0.09 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr5_+_49962772 | 0.09 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_135818979 | 0.09 |
ENST00000421378.2
ENST00000579057.1 ENST00000436554.1 ENST00000438618.2 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr15_+_43885252 | 0.08 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_+_27046988 | 0.08 |
ENST00000496182.1
|
RPL23A
|
ribosomal protein L23a |
| chr2_+_66918558 | 0.08 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr6_+_155537771 | 0.08 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr16_+_57139933 | 0.08 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr3_-_111852061 | 0.08 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr9_+_129097479 | 0.08 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr12_-_122658746 | 0.08 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr16_-_3306587 | 0.08 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chrX_+_100663243 | 0.08 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr1_+_38022513 | 0.08 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr12_-_12837423 | 0.08 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr1_-_161337662 | 0.08 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr15_-_52043722 | 0.08 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr1_+_162602244 | 0.08 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr7_+_108210012 | 0.07 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr8_-_74884399 | 0.07 |
ENST00000520210.1
ENST00000602840.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr13_-_49975632 | 0.07 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr13_+_92050928 | 0.07 |
ENST00000377067.3
|
GPC5
|
glypican 5 |
| chr17_+_27047244 | 0.07 |
ENST00000394938.4
ENST00000394935.3 ENST00000355731.4 |
RPL23A
|
ribosomal protein L23a |
| chr15_+_43985084 | 0.07 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr17_+_30814707 | 0.07 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr17_-_15522826 | 0.07 |
ENST00000395906.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr4_+_75230853 | 0.07 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr7_-_91794590 | 0.07 |
ENST00000430130.2
ENST00000454089.2 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr6_+_43739697 | 0.07 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr1_+_186265399 | 0.07 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr15_+_49715293 | 0.07 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr3_+_9944492 | 0.07 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr11_+_18417813 | 0.07 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chr9_-_16705069 | 0.07 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr1_+_179712298 | 0.07 |
ENST00000341785.4
|
FAM163A
|
family with sequence similarity 163, member A |
| chr17_-_38256973 | 0.07 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr2_-_68290106 | 0.07 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr1_+_36038971 | 0.07 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr10_+_71561630 | 0.07 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_-_159065741 | 0.07 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr15_+_63188009 | 0.07 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chrX_+_18725758 | 0.07 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr12_-_76425368 | 0.07 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr3_-_27764190 | 0.07 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr15_+_32933866 | 0.06 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr5_+_66124590 | 0.06 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_218683438 | 0.06 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr14_+_62164340 | 0.06 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr18_-_24237339 | 0.06 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr12_-_21910775 | 0.06 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr1_-_115053781 | 0.06 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr1_+_118148556 | 0.06 |
ENST00000369448.3
|
FAM46C
|
family with sequence similarity 46, member C |
| chr12_+_10331605 | 0.06 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr3_+_193853927 | 0.06 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr14_-_92413353 | 0.06 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr12_-_123756781 | 0.06 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr18_-_48346415 | 0.06 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr12_-_121973974 | 0.06 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr7_+_22766766 | 0.06 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr7_-_150754935 | 0.06 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr15_-_50647347 | 0.06 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr7_-_127032114 | 0.06 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr2_-_21266816 | 0.06 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr15_-_50647274 | 0.06 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr21_+_17566643 | 0.06 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_-_156685841 | 0.06 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr10_+_25305524 | 0.06 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr11_-_2924720 | 0.06 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr6_+_22221010 | 0.06 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr17_+_16120512 | 0.06 |
ENST00000581006.1
ENST00000584797.1 ENST00000498772.2 ENST00000225609.5 ENST00000395844.4 ENST00000477745.1 |
PIGL
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr2_-_190927447 | 0.06 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr19_+_7599128 | 0.06 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr9_+_137987825 | 0.06 |
ENST00000545657.1
|
OLFM1
|
olfactomedin 1 |
| chr12_+_52450298 | 0.06 |
ENST00000550582.2
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr14_-_36988882 | 0.06 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr11_-_88070920 | 0.06 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr12_+_54402790 | 0.06 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr3_-_124839648 | 0.06 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr5_-_159846066 | 0.06 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr3_+_140947563 | 0.06 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr16_-_10868853 | 0.06 |
ENST00000572428.1
|
TVP23A
|
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
| chr7_+_140373619 | 0.06 |
ENST00000483369.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr12_-_50616382 | 0.06 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr5_+_155753745 | 0.06 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr13_+_97874574 | 0.06 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr8_-_80680078 | 0.06 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr6_+_86159821 | 0.05 |
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr12_+_75874580 | 0.05 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr12_-_49581152 | 0.05 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chrX_-_102943022 | 0.05 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr5_-_87969130 | 0.05 |
ENST00000505030.1
ENST00000500197.2 |
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr2_-_207082748 | 0.05 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr4_+_100737954 | 0.05 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr3_+_25469724 | 0.05 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr11_-_62457371 | 0.05 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr15_+_45028719 | 0.05 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr5_+_135394840 | 0.05 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr16_-_85969774 | 0.05 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr1_+_244816237 | 0.05 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr18_-_47376197 | 0.05 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr20_-_14318248 | 0.05 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr21_-_35899113 | 0.05 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr2_+_42104692 | 0.05 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr8_-_97173020 | 0.05 |
ENST00000287020.5
|
GDF6
|
growth differentiation factor 6 |
| chrX_-_135056106 | 0.05 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr3_+_88188254 | 0.05 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr11_+_114271314 | 0.05 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7 |
| chr20_+_36149602 | 0.05 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr19_-_10445399 | 0.05 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr8_-_99954788 | 0.05 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr5_-_94620239 | 0.05 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr19_-_36523529 | 0.05 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr15_+_74466744 | 0.05 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr4_-_38806404 | 0.05 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr2_+_27237615 | 0.05 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr17_-_49337392 | 0.05 |
ENST00000376381.2
ENST00000586178.1 |
MBTD1
|
mbt domain containing 1 |
| chr9_-_73483926 | 0.05 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_+_114271251 | 0.05 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr1_-_156675564 | 0.05 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr8_+_94712895 | 0.05 |
ENST00000523453.1
ENST00000520955.1 |
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr19_+_40477062 | 0.05 |
ENST00000455878.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr17_+_4853442 | 0.05 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr9_-_127710292 | 0.05 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr18_-_3845321 | 0.05 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr12_-_122240792 | 0.05 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr8_+_94712732 | 0.05 |
ENST00000518322.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr7_+_112262421 | 0.05 |
ENST00000453459.1
|
AC002463.3
|
AC002463.3 |
| chr15_-_37392086 | 0.05 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chrX_-_37706815 | 0.05 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr4_+_80584903 | 0.05 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr8_-_38854332 | 0.05 |
ENST00000520152.1
ENST00000522142.1 |
TM2D2
|
TM2 domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.0 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.0 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.0 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.0 | GO:1903217 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.0 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.0 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |