Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
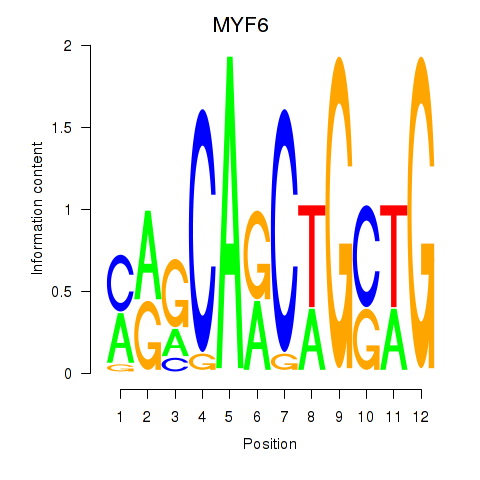
Results for MYF6
Z-value: 0.97
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | myogenic factor 6 |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_21905120 | 1.69 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr19_-_47734448 | 0.66 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr10_-_90967063 | 0.57 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr1_+_38273988 | 0.49 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr1_+_155583012 | 0.48 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_+_38273818 | 0.46 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr4_+_2043689 | 0.45 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr4_+_89300158 | 0.43 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr11_-_117747327 | 0.42 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr10_-_79397740 | 0.40 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr19_+_18451391 | 0.40 |
ENST00000269919.6
ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr4_+_2043777 | 0.39 |
ENST00000409860.1
|
C4orf48
|
chromosome 4 open reading frame 48 |
| chr19_-_18391708 | 0.39 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr6_+_31514622 | 0.37 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr3_-_196987309 | 0.37 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr16_+_691792 | 0.37 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr15_+_90744745 | 0.35 |
ENST00000558051.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_-_149783914 | 0.35 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr12_-_14967095 | 0.34 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr2_-_220408260 | 0.33 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr15_-_94614049 | 0.32 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr7_-_74867509 | 0.32 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr10_-_79397547 | 0.32 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_-_87367879 | 0.32 |
ENST00000568879.1
|
RP11-178L8.4
|
RP11-178L8.4 |
| chr17_+_39411636 | 0.32 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr5_-_158757895 | 0.31 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr1_-_98511756 | 0.31 |
ENST00000602984.1
ENST00000602852.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr2_-_220408430 | 0.30 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr19_+_13988061 | 0.29 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr18_-_53253112 | 0.29 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr6_+_33378517 | 0.27 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr4_+_102268904 | 0.27 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr17_-_80009650 | 0.27 |
ENST00000310496.4
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_+_183967409 | 0.26 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr7_+_100199800 | 0.26 |
ENST00000223061.5
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chrX_+_48681768 | 0.25 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr11_-_414948 | 0.24 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chrX_-_38080077 | 0.24 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr20_-_60942361 | 0.24 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr17_-_39538550 | 0.24 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr7_+_100860949 | 0.24 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr18_-_53253000 | 0.24 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr3_-_178789220 | 0.23 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr12_+_56137064 | 0.23 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr3_-_87040259 | 0.23 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr22_-_50964849 | 0.23 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_-_51014460 | 0.22 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr22_-_24989014 | 0.22 |
ENST00000318753.8
|
FAM211B
|
family with sequence similarity 211, member B |
| chrX_+_23685653 | 0.22 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr22_+_18893736 | 0.22 |
ENST00000331444.6
|
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr9_-_131940526 | 0.22 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr9_+_35042205 | 0.21 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr11_-_67120974 | 0.21 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr16_-_31076332 | 0.21 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr7_+_2559399 | 0.21 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_180165672 | 0.21 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr17_-_42452063 | 0.21 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr19_+_41699103 | 0.21 |
ENST00000597754.1
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr11_+_34999328 | 0.21 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr1_+_77997785 | 0.21 |
ENST00000478255.1
|
AK5
|
adenylate kinase 5 |
| chr22_-_30783075 | 0.21 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr6_+_157099036 | 0.21 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr16_-_88878305 | 0.21 |
ENST00000569616.1
ENST00000563655.1 ENST00000567713.1 ENST00000426324.2 ENST00000378364.3 |
APRT
|
adenine phosphoribosyltransferase |
| chr7_+_72848092 | 0.21 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr18_-_53253323 | 0.21 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr17_+_80009741 | 0.21 |
ENST00000578552.1
ENST00000320548.4 ENST00000581578.1 ENST00000583885.1 ENST00000583641.1 ENST00000581418.1 ENST00000355130.2 ENST00000306823.6 ENST00000392358.2 |
GPS1
|
G protein pathway suppressor 1 |
| chr16_-_776431 | 0.20 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr9_+_84304628 | 0.20 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr12_-_76425368 | 0.20 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr15_+_96873921 | 0.20 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chrX_+_46433193 | 0.20 |
ENST00000276055.3
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr19_-_51014588 | 0.20 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr2_+_217498105 | 0.20 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr11_-_65325664 | 0.20 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr11_-_62323702 | 0.20 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr19_+_54695098 | 0.20 |
ENST00000396388.2
|
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr1_+_180123969 | 0.20 |
ENST00000367602.3
ENST00000367600.5 |
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr17_-_42100474 | 0.19 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr1_+_55505184 | 0.19 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr1_-_201438282 | 0.19 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr21_+_46875424 | 0.19 |
ENST00000359759.4
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr11_-_14913765 | 0.19 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr1_-_27226928 | 0.19 |
ENST00000361720.5
|
GPATCH3
|
G patch domain containing 3 |
| chr11_-_117698787 | 0.19 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr6_+_37137939 | 0.19 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
| chr21_+_46875395 | 0.19 |
ENST00000355480.5
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr1_+_36024107 | 0.19 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr19_-_40730820 | 0.19 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr10_+_129845670 | 0.19 |
ENST00000467366.1
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr10_-_103603523 | 0.19 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr22_-_30783356 | 0.18 |
ENST00000382363.3
|
RNF215
|
ring finger protein 215 |
| chr6_-_33385854 | 0.18 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_-_79633590 | 0.18 |
ENST00000374741.3
ENST00000571503.1 |
OXLD1
|
oxidoreductase-like domain containing 1 |
| chr6_-_31514333 | 0.18 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr1_-_226076843 | 0.18 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr22_+_31199037 | 0.18 |
ENST00000424224.1
|
OSBP2
|
oxysterol binding protein 2 |
| chr22_-_20307532 | 0.18 |
ENST00000405465.3
ENST00000248879.3 |
DGCR6L
|
DiGeorge syndrome critical region gene 6-like |
| chr22_-_50964558 | 0.18 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr1_+_109792641 | 0.18 |
ENST00000271332.3
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr1_-_72748417 | 0.18 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr3_-_183966717 | 0.18 |
ENST00000446569.1
ENST00000418734.2 ENST00000397676.3 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr12_-_49318715 | 0.18 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr8_+_31497271 | 0.18 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr15_-_76352069 | 0.18 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr17_+_74261277 | 0.18 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr4_-_140223614 | 0.18 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_+_20915409 | 0.18 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr12_+_52445191 | 0.18 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr8_-_123706338 | 0.18 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr11_+_114168773 | 0.18 |
ENST00000542647.1
ENST00000545255.1 |
NNMT
|
nicotinamide N-methyltransferase |
| chr16_-_31076273 | 0.17 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr16_-_4852616 | 0.17 |
ENST00000591392.1
ENST00000587711.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr1_-_9189144 | 0.17 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr20_+_4129496 | 0.17 |
ENST00000346595.2
|
SMOX
|
spermine oxidase |
| chr16_-_29910365 | 0.17 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr20_+_62367989 | 0.17 |
ENST00000309546.3
|
LIME1
|
Lck interacting transmembrane adaptor 1 |
| chr11_+_71939512 | 0.17 |
ENST00000540329.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr17_+_42634844 | 0.17 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr12_+_122241928 | 0.17 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr16_+_30935896 | 0.17 |
ENST00000562319.1
ENST00000380310.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr7_-_102283238 | 0.17 |
ENST00000340457.8
|
UPK3BL
|
uroplakin 3B-like |
| chr1_-_57045228 | 0.16 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr5_-_72744336 | 0.16 |
ENST00000499003.3
|
FOXD1
|
forkhead box D1 |
| chr1_-_6662919 | 0.16 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr11_-_9113137 | 0.16 |
ENST00000520467.1
ENST00000309263.3 ENST00000457346.2 |
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr22_-_42342692 | 0.16 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr1_+_161284047 | 0.16 |
ENST00000367975.2
ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr19_+_49128209 | 0.16 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr4_+_89299885 | 0.16 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_+_41103063 | 0.16 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_-_7760457 | 0.16 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr18_+_19750894 | 0.16 |
ENST00000581694.1
|
GATA6
|
GATA binding protein 6 |
| chr17_+_39421591 | 0.16 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr15_+_59908633 | 0.16 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr17_+_39405939 | 0.16 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr17_-_41132088 | 0.16 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr11_+_77300669 | 0.16 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr8_+_145438870 | 0.15 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr16_+_2285817 | 0.15 |
ENST00000564065.1
|
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr20_-_23731569 | 0.15 |
ENST00000304749.2
|
CST1
|
cystatin SN |
| chr5_+_174151536 | 0.15 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr19_-_3600549 | 0.15 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr14_-_54423529 | 0.15 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr8_-_22014255 | 0.15 |
ENST00000424267.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr14_+_73706308 | 0.15 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr20_-_60942326 | 0.15 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr17_+_42429493 | 0.15 |
ENST00000586242.1
|
GRN
|
granulin |
| chr10_+_51748078 | 0.15 |
ENST00000374056.4
|
AGAP6
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 6 |
| chr6_-_31940065 | 0.15 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr11_+_66115304 | 0.15 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr1_+_10509971 | 0.15 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr2_+_74212073 | 0.15 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr9_-_35619539 | 0.15 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr11_+_22688150 | 0.15 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr19_-_376011 | 0.14 |
ENST00000342640.4
|
THEG
|
theg spermatid protein |
| chr7_-_100171270 | 0.14 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr17_-_43138463 | 0.14 |
ENST00000310604.4
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr5_+_140213815 | 0.14 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chrX_+_153060090 | 0.14 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr19_-_19774473 | 0.14 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr3_+_49059038 | 0.14 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr11_-_66115032 | 0.14 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr2_+_202937972 | 0.14 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr16_+_5121814 | 0.14 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chrX_-_153285251 | 0.14 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr22_+_29702572 | 0.14 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr17_+_74261413 | 0.14 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr19_+_18451439 | 0.14 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr1_-_1535455 | 0.14 |
ENST00000422725.1
|
C1orf233
|
chromosome 1 open reading frame 233 |
| chr7_-_27183263 | 0.14 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr7_+_100210133 | 0.14 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr17_-_79166176 | 0.13 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr6_+_100054606 | 0.13 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr1_-_63782888 | 0.13 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr15_+_79603404 | 0.13 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chr3_-_183967296 | 0.13 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chrX_-_135849484 | 0.13 |
ENST00000370620.1
ENST00000535227.1 |
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr12_+_56521840 | 0.13 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr3_-_49058479 | 0.13 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr12_-_92539614 | 0.13 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr11_-_65325430 | 0.13 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr1_+_209941942 | 0.13 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr13_+_36050881 | 0.13 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr8_-_23712312 | 0.13 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr7_-_137028534 | 0.13 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr12_-_42632127 | 0.13 |
ENST00000555248.2
|
YAF2
|
YY1 associated factor 2 |
| chr1_+_51434357 | 0.13 |
ENST00000396148.1
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr19_-_40324767 | 0.13 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr11_-_117698765 | 0.13 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chrX_-_100641155 | 0.13 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr1_-_1009683 | 0.13 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr5_-_55777586 | 0.13 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr1_-_20987851 | 0.13 |
ENST00000464364.1
ENST00000602624.2 |
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr10_+_102758105 | 0.13 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr19_+_54371114 | 0.13 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr3_-_129325660 | 0.13 |
ENST00000324093.4
ENST00000393239.1 |
PLXND1
|
plexin D1 |
| chr5_-_137610254 | 0.13 |
ENST00000378362.3
|
GFRA3
|
GDNF family receptor alpha 3 |
| chr6_-_29600559 | 0.13 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr17_+_73512594 | 0.13 |
ENST00000333213.6
|
TSEN54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr19_+_13875316 | 0.12 |
ENST00000319545.8
ENST00000593245.1 ENST00000040663.6 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.5 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 0.2 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.2 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.2 | GO:0048390 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) |
| 0.0 | 0.5 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.3 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.1 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.0 | 0.1 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:0060738 | positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) sclerotome development(GO:0061056) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.0 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 0.0 | 0.0 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 0.0 | 0.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0003264 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.6 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:1902995 | positive regulation of engulfment of apoptotic cell(GO:1901076) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.0 | GO:0002583 | regulation of antigen processing and presentation of peptide antigen(GO:0002583) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.0 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.0 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.4 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |