Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
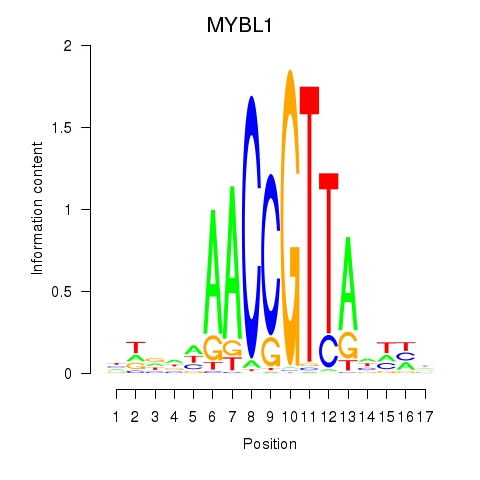
Results for MYBL1
Z-value: 0.34
Transcription factors associated with MYBL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL1
|
ENSG00000185697.12 | MYB proto-oncogene like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL1 | hg19_v2_chr8_-_67525473_67525518 | 0.33 | 6.7e-01 | Click! |
Activity profile of MYBL1 motif
Sorted Z-values of MYBL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_36239576 | 0.22 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr6_+_27782788 | 0.14 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr16_+_85832146 | 0.13 |
ENST00000565078.1
|
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr22_+_43011247 | 0.12 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr11_+_29181503 | 0.12 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr16_-_25122735 | 0.11 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr21_-_43816152 | 0.11 |
ENST00000433957.2
ENST00000398397.3 |
TMPRSS3
|
transmembrane protease, serine 3 |
| chr19_-_6199521 | 0.10 |
ENST00000592883.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_-_34978980 | 0.10 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr18_-_71815051 | 0.10 |
ENST00000582526.1
ENST00000419743.2 |
FBXO15
|
F-box protein 15 |
| chr4_-_6675550 | 0.10 |
ENST00000513179.1
ENST00000515205.1 |
RP11-539L10.3
|
RP11-539L10.3 |
| chr19_-_6110555 | 0.10 |
ENST00000593241.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr6_-_133119668 | 0.09 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr9_+_6716478 | 0.08 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr19_-_52552051 | 0.08 |
ENST00000221315.5
|
ZNF432
|
zinc finger protein 432 |
| chr14_+_45366518 | 0.08 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr18_+_3247779 | 0.08 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_-_57081940 | 0.08 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr13_-_49987885 | 0.07 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chrX_-_140673133 | 0.07 |
ENST00000370519.3
|
SPANXA1
|
sperm protein associated with the nucleus, X-linked, family member A1 |
| chrX_+_71996972 | 0.07 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr12_+_107349497 | 0.07 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr12_-_76478446 | 0.07 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_+_7053228 | 0.07 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr6_-_27782548 | 0.07 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr17_-_7835228 | 0.07 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr20_+_34287194 | 0.07 |
ENST00000374078.1
ENST00000374077.3 |
ROMO1
|
reactive oxygen species modulator 1 |
| chrX_-_46404811 | 0.07 |
ENST00000518708.1
ENST00000523374.1 |
ZNF674
|
zinc finger protein 674 |
| chr12_-_14996355 | 0.06 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr4_+_11470867 | 0.06 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr17_+_16284749 | 0.06 |
ENST00000578706.1
|
UBB
|
ubiquitin B |
| chrX_+_46404928 | 0.06 |
ENST00000421685.2
ENST00000609887.1 |
ZNF674-AS1
|
ZNF674 antisense RNA 1 (head to head) |
| chr6_+_161123270 | 0.06 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr16_+_56642489 | 0.06 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr19_+_1495362 | 0.06 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr19_-_52551814 | 0.06 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr1_+_92414952 | 0.06 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr19_+_38826415 | 0.06 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr10_-_91174215 | 0.06 |
ENST00000371837.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr8_+_95565947 | 0.06 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr10_-_28623368 | 0.06 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_-_214148921 | 0.06 |
ENST00000360083.3
|
AC079610.2
|
AC079610.2 |
| chr6_+_13272904 | 0.06 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr19_+_37709076 | 0.06 |
ENST00000590503.1
ENST00000589413.1 |
ZNF383
|
zinc finger protein 383 |
| chr12_+_25055243 | 0.06 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr4_+_103790462 | 0.06 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr18_-_70211691 | 0.06 |
ENST00000269503.4
|
CBLN2
|
cerebellin 2 precursor |
| chr5_-_88179017 | 0.06 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr7_-_1980128 | 0.05 |
ENST00000437877.1
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr4_-_80994619 | 0.05 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr17_+_16284104 | 0.05 |
ENST00000577958.1
ENST00000302182.3 ENST00000577640.1 |
UBB
|
ubiquitin B |
| chr4_-_147442982 | 0.05 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr11_+_47600562 | 0.05 |
ENST00000263774.4
ENST00000529276.1 ENST00000528192.1 ENST00000530295.1 ENST00000534208.1 ENST00000534716.2 |
NDUFS3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| chr12_-_76478686 | 0.05 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr16_+_4784273 | 0.05 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr2_-_86564740 | 0.05 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr15_-_30261066 | 0.05 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr12_-_62997214 | 0.05 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr6_-_133119649 | 0.05 |
ENST00000367918.1
|
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr3_-_169482840 | 0.05 |
ENST00000602385.1
|
TERC
|
telomerase RNA component |
| chr4_-_120988229 | 0.05 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr3_+_32737027 | 0.05 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr12_+_7053172 | 0.05 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr1_+_40974431 | 0.05 |
ENST00000296380.4
ENST00000432259.1 ENST00000418186.1 |
EXO5
|
exonuclease 5 |
| chr7_+_7222233 | 0.05 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr19_-_38183201 | 0.05 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr1_+_2066252 | 0.05 |
ENST00000466352.1
|
PRKCZ
|
protein kinase C, zeta |
| chr1_-_43855444 | 0.05 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr17_+_16284604 | 0.05 |
ENST00000395839.1
ENST00000395837.1 |
UBB
|
ubiquitin B |
| chr6_+_89855765 | 0.05 |
ENST00000275072.4
|
PM20D2
|
peptidase M20 domain containing 2 |
| chr5_-_74162739 | 0.05 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr12_-_123201337 | 0.05 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr17_+_41363854 | 0.05 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr19_+_57791419 | 0.05 |
ENST00000537645.1
|
ZNF460
|
zinc finger protein 460 |
| chr1_+_172502336 | 0.05 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr7_-_76255444 | 0.05 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr4_-_4544061 | 0.05 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr1_-_43855479 | 0.04 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr22_+_32455111 | 0.04 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr11_-_9336234 | 0.04 |
ENST00000528080.1
|
TMEM41B
|
transmembrane protein 41B |
| chr2_-_37501692 | 0.04 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr3_+_186501979 | 0.04 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr10_+_90562487 | 0.04 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr6_+_71377567 | 0.04 |
ENST00000422334.2
|
SMAP1
|
small ArfGAP 1 |
| chr10_+_90562705 | 0.04 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr8_+_101162812 | 0.04 |
ENST00000353107.3
ENST00000522439.1 |
POLR2K
|
polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa |
| chr17_-_56769382 | 0.04 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr4_+_166248775 | 0.04 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr11_+_33037652 | 0.04 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr1_-_45805752 | 0.04 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr17_-_30668887 | 0.04 |
ENST00000581747.1
ENST00000583334.1 ENST00000580558.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr3_-_134204815 | 0.04 |
ENST00000514612.1
ENST00000510994.1 ENST00000354910.5 |
ANAPC13
|
anaphase promoting complex subunit 13 |
| chr4_-_147443043 | 0.04 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr5_-_147211190 | 0.04 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr8_+_104310661 | 0.04 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr11_+_20409070 | 0.04 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr4_+_152020736 | 0.04 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr11_-_118927735 | 0.04 |
ENST00000526656.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr12_+_69753448 | 0.04 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr17_+_48133330 | 0.04 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr9_-_34126730 | 0.04 |
ENST00000361264.4
|
DCAF12
|
DDB1 and CUL4 associated factor 12 |
| chr8_+_96037205 | 0.04 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr16_-_2205352 | 0.04 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr15_-_79103757 | 0.04 |
ENST00000388820.4
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr3_+_134204551 | 0.04 |
ENST00000332047.5
ENST00000354446.3 |
CEP63
|
centrosomal protein 63kDa |
| chr3_+_49977523 | 0.04 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr1_+_163291680 | 0.04 |
ENST00000450453.2
ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr20_+_52824367 | 0.03 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr14_+_45366472 | 0.03 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr2_-_70781087 | 0.03 |
ENST00000394241.3
ENST00000295400.6 |
TGFA
|
transforming growth factor, alpha |
| chr3_+_49977490 | 0.03 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr11_+_118938485 | 0.03 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr10_+_71561630 | 0.03 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_-_43708378 | 0.03 |
ENST00000599746.1
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr1_-_39395165 | 0.03 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr18_+_3247413 | 0.03 |
ENST00000579226.1
ENST00000217652.3 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr7_-_2272566 | 0.03 |
ENST00000402746.1
ENST00000265854.7 ENST00000429779.1 ENST00000399654.2 |
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr6_+_142468361 | 0.03 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr15_+_66797455 | 0.03 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr8_-_23315131 | 0.03 |
ENST00000518718.1
|
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr20_+_34802295 | 0.03 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr6_-_134639180 | 0.03 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_19615744 | 0.03 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr18_-_53735601 | 0.03 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr2_+_66918558 | 0.03 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr11_-_3013497 | 0.03 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr8_+_96037255 | 0.03 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr10_-_14613968 | 0.03 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr10_+_76969909 | 0.03 |
ENST00000298468.5
ENST00000543351.1 |
VDAC2
|
voltage-dependent anion channel 2 |
| chr8_-_145652336 | 0.03 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr15_+_38746307 | 0.03 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr19_+_18682661 | 0.03 |
ENST00000596273.1
ENST00000442744.2 ENST00000595683.1 ENST00000599256.1 ENST00000595158.1 ENST00000598780.1 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr6_+_122793058 | 0.03 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr11_+_93479588 | 0.03 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr2_+_219472637 | 0.03 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr19_-_9903666 | 0.03 |
ENST00000592587.1
|
ZNF846
|
zinc finger protein 846 |
| chr16_+_524900 | 0.03 |
ENST00000449879.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr3_-_122746628 | 0.03 |
ENST00000357599.3
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr4_+_174818390 | 0.03 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr12_+_107349606 | 0.03 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr6_+_142468383 | 0.03 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr20_+_18488528 | 0.03 |
ENST00000377465.1
|
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr16_+_29802036 | 0.03 |
ENST00000561482.1
ENST00000160827.4 ENST00000569636.2 ENST00000400750.2 |
KIF22
|
kinesin family member 22 |
| chr18_+_20513782 | 0.03 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr3_-_192445289 | 0.03 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr12_-_104359475 | 0.03 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr19_-_53360853 | 0.03 |
ENST00000596559.1
ENST00000594602.1 ENST00000595646.1 ENST00000597924.1 ENST00000396409.4 ENST00000390651.4 ENST00000243639.4 |
ZNF28
ZNF468
|
zinc finger protein 28 zinc finger protein 468 |
| chr7_+_23221438 | 0.03 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chrM_+_4431 | 0.03 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_118927880 | 0.03 |
ENST00000527038.2
|
HYOU1
|
hypoxia up-regulated 1 |
| chr19_-_14640005 | 0.03 |
ENST00000596853.1
ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_-_124981475 | 0.03 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr10_-_35379524 | 0.03 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr7_-_148581360 | 0.03 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr4_-_69083720 | 0.03 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr8_-_81083731 | 0.03 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr1_-_1510204 | 0.03 |
ENST00000291386.3
|
SSU72
|
SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) |
| chr11_+_122753391 | 0.03 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr6_+_11094266 | 0.03 |
ENST00000416247.2
|
SMIM13
|
small integral membrane protein 13 |
| chr5_+_139055021 | 0.03 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr21_-_15755446 | 0.03 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chr18_-_48346415 | 0.03 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr8_+_124084899 | 0.03 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr2_+_172864490 | 0.03 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr19_+_40502938 | 0.02 |
ENST00000599504.1
ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546
|
zinc finger protein 546 |
| chr2_+_202098203 | 0.02 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr5_+_140213815 | 0.02 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr5_-_40835303 | 0.02 |
ENST00000509877.1
ENST00000508493.1 ENST00000274242.5 |
RPL37
|
ribosomal protein L37 |
| chr17_+_48243352 | 0.02 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr6_-_138539627 | 0.02 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr3_+_12598563 | 0.02 |
ENST00000411987.1
ENST00000448482.1 |
MKRN2
|
makorin ring finger protein 2 |
| chr6_+_71377473 | 0.02 |
ENST00000370452.3
ENST00000316999.5 ENST00000370455.3 |
SMAP1
|
small ArfGAP 1 |
| chr19_+_21324827 | 0.02 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr3_-_50336278 | 0.02 |
ENST00000359051.3
ENST00000417393.1 ENST00000442620.1 ENST00000452674.1 |
HYAL3
NAT6
|
hyaluronoglucosaminidase 3 N-acetyltransferase 6 (GCN5-related) |
| chr1_+_210406121 | 0.02 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr3_-_51975942 | 0.02 |
ENST00000232888.6
|
RRP9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr1_+_162039558 | 0.02 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr17_-_30669138 | 0.02 |
ENST00000225805.4
ENST00000577809.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr1_+_99127265 | 0.02 |
ENST00000306121.3
|
SNX7
|
sorting nexin 7 |
| chr19_-_55919087 | 0.02 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr19_+_58111241 | 0.02 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr12_-_57082060 | 0.02 |
ENST00000448157.2
ENST00000414274.3 ENST00000262033.6 ENST00000456859.2 |
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr10_-_82116497 | 0.02 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr19_-_40023450 | 0.02 |
ENST00000326282.4
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr3_+_73045936 | 0.02 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr9_+_71394945 | 0.02 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr17_+_73201754 | 0.02 |
ENST00000583569.1
ENST00000245544.4 ENST00000579324.1 ENST00000541827.1 ENST00000579298.1 ENST00000447371.2 |
NUP85
|
nucleoporin 85kDa |
| chr5_+_140593509 | 0.02 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr8_-_25315905 | 0.02 |
ENST00000221200.4
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr21_-_33984456 | 0.02 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr1_+_180601139 | 0.02 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr1_-_153581798 | 0.02 |
ENST00000368704.1
ENST00000368705.2 |
S100A16
|
S100 calcium binding protein A16 |
| chr7_+_99102573 | 0.02 |
ENST00000394170.2
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr6_-_31510181 | 0.02 |
ENST00000458640.1
ENST00000396172.1 ENST00000417556.2 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr1_+_53662101 | 0.02 |
ENST00000371486.3
|
CPT2
|
carnitine palmitoyltransferase 2 |
| chr7_+_73106926 | 0.02 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr5_+_10250328 | 0.02 |
ENST00000515390.1
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr13_-_21635631 | 0.02 |
ENST00000382592.4
|
LATS2
|
large tumor suppressor kinase 2 |
| chr14_-_78174344 | 0.02 |
ENST00000216489.3
|
ALKBH1
|
alkB, alkylation repair homolog 1 (E. coli) |
| chr19_+_12780512 | 0.02 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.1 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.0 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |