Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
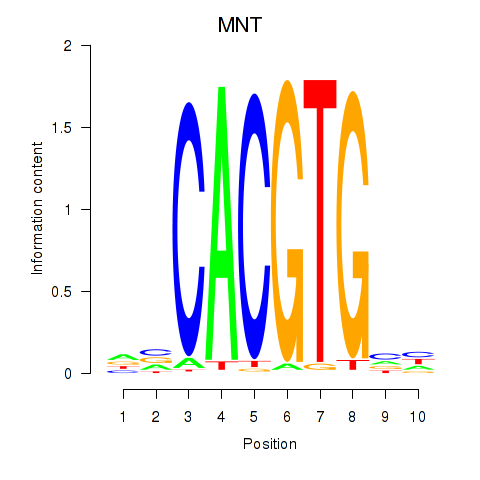
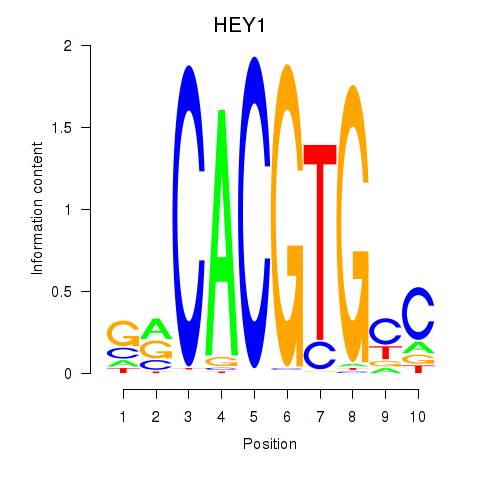
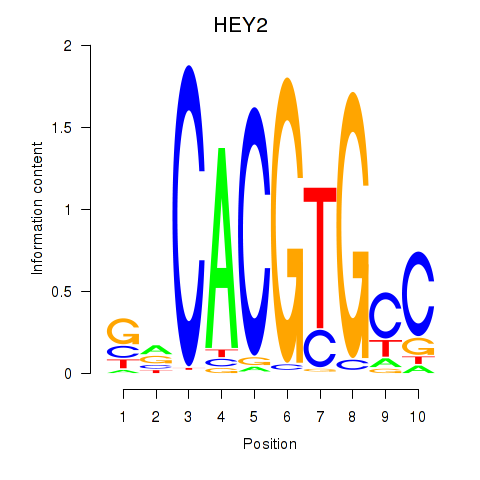
Results for MNT_HEY1_HEY2
Z-value: 0.70
Transcription factors associated with MNT_HEY1_HEY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MNT
|
ENSG00000070444.10 | MAX network transcriptional repressor |
|
HEY1
|
ENSG00000164683.12 | hes related family bHLH transcription factor with YRPW motif 1 |
|
HEY2
|
ENSG00000135547.4 | hes related family bHLH transcription factor with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HEY2 | hg19_v2_chr6_+_126070726_126070768 | 0.48 | 5.2e-01 | Click! |
| HEY1 | hg19_v2_chr8_-_80680078_80680101 | 0.34 | 6.6e-01 | Click! |
| MNT | hg19_v2_chr17_-_2304365_2304412 | 0.22 | 7.8e-01 | Click! |
Activity profile of MNT_HEY1_HEY2 motif
Sorted Z-values of MNT_HEY1_HEY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_112451222 | 0.79 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr2_-_232328867 | 0.67 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr20_-_3219359 | 0.63 |
ENST00000437836.2
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr3_+_186648507 | 0.50 |
ENST00000458216.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr15_+_44084040 | 0.50 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr4_-_1107306 | 0.47 |
ENST00000433731.2
ENST00000333673.5 ENST00000382968.5 |
RNF212
|
ring finger protein 212 |
| chr19_+_7587555 | 0.43 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr19_+_1261106 | 0.43 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr15_+_44084503 | 0.43 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr17_-_6915646 | 0.37 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr16_+_50280020 | 0.37 |
ENST00000564965.1
|
ADCY7
|
adenylate cyclase 7 |
| chr15_+_82555125 | 0.35 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr12_+_64845864 | 0.33 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr6_-_163834852 | 0.32 |
ENST00000604200.1
|
CAHM
|
colon adenocarcinoma hypermethylated (non-protein coding) |
| chr9_-_100684769 | 0.31 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr2_+_232575168 | 0.29 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr19_+_5681153 | 0.29 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr7_-_155089251 | 0.29 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr1_+_154193643 | 0.28 |
ENST00000456325.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr1_+_150254936 | 0.28 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr14_-_54955376 | 0.28 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr5_-_133706695 | 0.28 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr16_-_4588469 | 0.25 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr10_-_46090334 | 0.25 |
ENST00000395771.3
ENST00000319836.3 |
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr11_-_69490135 | 0.25 |
ENST00000542341.1
|
ORAOV1
|
oral cancer overexpressed 1 |
| chr12_-_9102224 | 0.24 |
ENST00000543845.1
ENST00000544245.1 |
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr7_-_130597935 | 0.24 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr3_-_52719912 | 0.24 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr2_+_232575128 | 0.23 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr6_-_151773232 | 0.23 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr10_-_46089939 | 0.23 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr10_+_46222648 | 0.22 |
ENST00000336378.4
ENST00000540872.1 ENST00000537517.1 ENST00000374362.2 ENST00000359860.4 ENST00000420848.1 |
FAM21C
|
family with sequence similarity 21, member C |
| chr2_+_30670209 | 0.22 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr1_-_26233423 | 0.21 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr10_-_6019455 | 0.21 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr1_+_154193325 | 0.21 |
ENST00000428931.1
ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L
|
ubiquitin associated protein 2-like |
| chr3_-_149688971 | 0.21 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr8_+_109455830 | 0.21 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr7_+_100464760 | 0.21 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr3_+_122785895 | 0.21 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr10_+_51827648 | 0.20 |
ENST00000351071.6
ENST00000314664.7 ENST00000282633.5 |
FAM21A
|
family with sequence similarity 21, member A |
| chr16_-_18937072 | 0.20 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr14_-_92572894 | 0.20 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr10_-_31320860 | 0.20 |
ENST00000436087.2
ENST00000442986.1 ENST00000413025.1 ENST00000452305.1 |
ZNF438
|
zinc finger protein 438 |
| chr12_+_95611536 | 0.20 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr2_+_121493717 | 0.20 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr12_-_6715808 | 0.20 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr5_-_176738883 | 0.20 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr7_-_27170352 | 0.19 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr1_-_26232951 | 0.19 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr5_-_178054014 | 0.19 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr7_+_23636992 | 0.19 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr4_-_17783135 | 0.19 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr1_-_111746966 | 0.19 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr4_+_129732419 | 0.19 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr22_-_29663690 | 0.18 |
ENST00000406335.1
|
RHBDD3
|
rhomboid domain containing 3 |
| chr12_+_64845660 | 0.18 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1 |
| chr9_-_100684845 | 0.18 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr10_-_31320840 | 0.18 |
ENST00000375311.1
|
ZNF438
|
zinc finger protein 438 |
| chr2_+_46926326 | 0.18 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr12_-_123380610 | 0.18 |
ENST00000535765.1
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr19_-_51142540 | 0.18 |
ENST00000598997.1
|
SYT3
|
synaptotagmin III |
| chrX_+_102469997 | 0.18 |
ENST00000372695.5
ENST00000372691.3 |
BEX4
|
brain expressed, X-linked 4 |
| chr6_-_90529418 | 0.17 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr10_+_180643 | 0.17 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr4_-_83295296 | 0.17 |
ENST00000507010.1
ENST00000503822.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr12_+_122356488 | 0.17 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr9_+_130159433 | 0.17 |
ENST00000451404.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr10_-_6019552 | 0.17 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr10_+_180405 | 0.16 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr12_+_9102632 | 0.16 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr11_-_116968987 | 0.16 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr1_+_11866270 | 0.16 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr9_-_135545380 | 0.16 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr1_-_110613276 | 0.16 |
ENST00000369792.4
|
ALX3
|
ALX homeobox 3 |
| chr11_+_65686728 | 0.16 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr5_-_150138246 | 0.16 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chr8_+_6566206 | 0.16 |
ENST00000518327.1
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr2_-_198364581 | 0.15 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr4_+_76649753 | 0.15 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr17_-_58469591 | 0.15 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr6_-_56112235 | 0.15 |
ENST00000370817.3
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr19_-_47164386 | 0.15 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_-_92465868 | 0.15 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr21_-_16374688 | 0.15 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr14_-_91884150 | 0.15 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr3_+_133292851 | 0.15 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr15_-_52971544 | 0.15 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr17_-_41465674 | 0.15 |
ENST00000592135.1
ENST00000587874.1 ENST00000588654.1 ENST00000592094.1 |
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr11_-_69490073 | 0.15 |
ENST00000535657.1
ENST00000539414.1 ENST00000536870.1 ENST00000538554.2 ENST00000279147.4 |
ORAOV1
|
oral cancer overexpressed 1 |
| chr16_-_54963026 | 0.14 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr2_+_173420697 | 0.14 |
ENST00000282077.3
ENST00000392571.2 ENST00000410055.1 |
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr16_+_88636875 | 0.14 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr8_+_38614754 | 0.14 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr2_+_240323439 | 0.14 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr2_+_198365122 | 0.14 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr1_-_180991978 | 0.14 |
ENST00000542060.1
ENST00000258301.5 |
STX6
|
syntaxin 6 |
| chr1_+_43637996 | 0.14 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr19_+_46144884 | 0.14 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr8_-_124553437 | 0.14 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr11_+_18720316 | 0.14 |
ENST00000280734.2
|
TMEM86A
|
transmembrane protein 86A |
| chr1_-_151319654 | 0.13 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr1_-_231376836 | 0.13 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr8_-_144679602 | 0.13 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr9_+_37120536 | 0.13 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr3_-_45883558 | 0.13 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr2_-_68384603 | 0.13 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr11_-_61684962 | 0.13 |
ENST00000394836.2
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr18_+_9136758 | 0.13 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr11_+_133902547 | 0.13 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr1_+_246729815 | 0.13 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr2_+_111878483 | 0.13 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr2_+_201171268 | 0.13 |
ENST00000423749.1
ENST00000428692.1 ENST00000457757.1 ENST00000453663.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr19_-_19739321 | 0.13 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr1_-_78149041 | 0.13 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chrX_+_16804544 | 0.13 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr3_-_150481218 | 0.13 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr11_+_72929402 | 0.12 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr22_+_29469100 | 0.12 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr17_-_17109579 | 0.12 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr7_-_150675372 | 0.12 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr9_+_96793076 | 0.12 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr7_-_42951509 | 0.12 |
ENST00000438029.1
ENST00000432637.1 ENST00000447342.1 ENST00000431882.2 ENST00000350427.4 ENST00000425683.1 |
C7orf25
|
chromosome 7 open reading frame 25 |
| chr19_+_1407733 | 0.12 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr7_-_27205136 | 0.12 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr2_-_176867534 | 0.12 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr1_-_222886526 | 0.12 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr3_-_52719810 | 0.12 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr19_+_50031547 | 0.12 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr9_+_37079968 | 0.12 |
ENST00000588403.1
|
RP11-220I1.1
|
RP11-220I1.1 |
| chr1_+_28261621 | 0.12 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_-_64013663 | 0.12 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr4_+_9446156 | 0.12 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr2_-_40006357 | 0.11 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr2_+_208576259 | 0.11 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr5_-_1799864 | 0.11 |
ENST00000510999.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr10_-_16859442 | 0.11 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr1_+_244998918 | 0.11 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr2_+_56411131 | 0.11 |
ENST00000407595.2
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr10_-_120840309 | 0.11 |
ENST00000369144.3
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A |
| chr1_+_45140360 | 0.11 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr11_+_72929319 | 0.11 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr11_+_75479850 | 0.11 |
ENST00000376262.3
ENST00000604733.1 |
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr8_-_144442136 | 0.11 |
ENST00000519148.1
|
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr16_-_4897266 | 0.11 |
ENST00000591451.1
ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr8_+_17354587 | 0.11 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr1_-_52520828 | 0.11 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr4_+_129732467 | 0.11 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr2_+_198365095 | 0.11 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr7_+_99070464 | 0.11 |
ENST00000331410.5
ENST00000483089.1 ENST00000448667.1 ENST00000493485.1 |
ZNF789
|
zinc finger protein 789 |
| chr4_+_164415855 | 0.11 |
ENST00000508268.1
|
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr6_+_73331918 | 0.11 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr8_-_144442094 | 0.11 |
ENST00000521193.1
ENST00000520950.1 |
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr9_+_129987488 | 0.11 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_+_113666748 | 0.11 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr15_+_41913690 | 0.11 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr12_-_122750957 | 0.11 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr1_-_26232522 | 0.11 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr4_+_183065793 | 0.11 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr3_+_52720187 | 0.11 |
ENST00000474423.1
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr18_+_9913977 | 0.11 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr19_+_49458107 | 0.11 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr1_+_109642799 | 0.11 |
ENST00000602755.1
|
SCARNA2
|
small Cajal body-specific RNA 2 |
| chrX_-_20134713 | 0.11 |
ENST00000452324.3
|
MAP7D2
|
MAP7 domain containing 2 |
| chr11_+_65686802 | 0.11 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr1_-_11866034 | 0.10 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr7_-_27169801 | 0.10 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chrX_+_38420783 | 0.10 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr4_-_146101304 | 0.10 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr2_+_187350973 | 0.10 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr9_-_35691017 | 0.10 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta) |
| chr6_+_33257427 | 0.10 |
ENST00000463584.1
|
PFDN6
|
prefoldin subunit 6 |
| chr4_-_83351294 | 0.10 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr9_-_135819987 | 0.10 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr17_+_48450575 | 0.10 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr14_-_39901618 | 0.10 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr2_-_40006289 | 0.10 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr21_+_45527171 | 0.10 |
ENST00000291576.7
ENST00000456705.1 |
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr16_-_81129845 | 0.10 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr9_+_37079888 | 0.10 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr19_-_36545128 | 0.10 |
ENST00000538849.1
|
THAP8
|
THAP domain containing 8 |
| chr6_+_33257346 | 0.10 |
ENST00000374606.5
ENST00000374610.2 ENST00000374607.1 |
PFDN6
|
prefoldin subunit 6 |
| chrX_+_55744228 | 0.10 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr2_+_233562015 | 0.10 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr10_-_104474128 | 0.10 |
ENST00000260746.5
|
ARL3
|
ADP-ribosylation factor-like 3 |
| chr12_+_104235229 | 0.10 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr7_-_32931623 | 0.10 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr19_-_46974741 | 0.10 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr8_+_142402089 | 0.10 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr19_-_13030071 | 0.10 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr4_-_103997455 | 0.10 |
ENST00000362026.3
ENST00000503103.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr17_+_27573875 | 0.10 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr12_-_6756559 | 0.10 |
ENST00000536350.1
ENST00000414226.2 ENST00000546114.1 |
ACRBP
|
acrosin binding protein |
| chr2_-_232329186 | 0.10 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chr13_-_52027134 | 0.10 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr20_-_61051026 | 0.10 |
ENST00000252997.2
|
GATA5
|
GATA binding protein 5 |
| chr8_+_120885949 | 0.10 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr5_-_142784101 | 0.10 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_-_131132658 | 0.10 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr8_+_109455845 | 0.10 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MNT_HEY1_HEY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 0.5 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.1 | 0.3 | GO:1990828 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.1 | 0.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.2 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.7 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0061461 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.0 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.6 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0044417 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:1902683 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1902074 | response to salt(GO:1902074) |
| 0.0 | 0.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.0 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.1 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.0 | GO:0048320 | inhibition of neuroepithelial cell differentiation(GO:0002085) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.0 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.0 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.0 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0046696 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.0 | GO:0009346 | citrate lyase complex(GO:0009346) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.6 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME CELL CYCLE | Genes involved in Cell Cycle |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |