Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
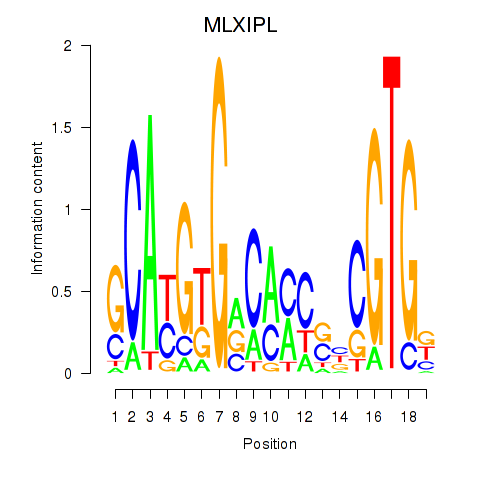
Results for MLXIPL
Z-value: 0.50
Transcription factors associated with MLXIPL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MLXIPL
|
ENSG00000009950.11 | MLX interacting protein like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MLXIPL | hg19_v2_chr7_-_73038822_73038862 | -0.74 | 2.6e-01 | Click! |
Activity profile of MLXIPL motif
Sorted Z-values of MLXIPL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_23145288 | 0.20 |
ENST00000419813.1
|
KLHL7-AS1
|
KLHL7 antisense RNA 1 (head to head) |
| chr19_+_23257988 | 0.20 |
ENST00000594318.1
ENST00000600223.1 ENST00000593635.1 |
CTD-2291D10.4
ZNF730
|
CTD-2291D10.4 zinc finger protein 730 |
| chr8_-_1922789 | 0.16 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr9_-_88896977 | 0.15 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr10_+_91461413 | 0.15 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr19_+_50031547 | 0.14 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr10_-_75634219 | 0.13 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr16_+_84225029 | 0.13 |
ENST00000567685.1
|
ADAD2
|
adenosine deaminase domain containing 2 |
| chr1_-_40237020 | 0.13 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr2_-_27851745 | 0.12 |
ENST00000394775.3
ENST00000522876.1 |
CCDC121
|
coiled-coil domain containing 121 |
| chr15_+_96897466 | 0.12 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr7_+_74072288 | 0.11 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr4_+_187112674 | 0.11 |
ENST00000378802.4
|
CYP4V2
|
cytochrome P450, family 4, subfamily V, polypeptide 2 |
| chr15_-_77712477 | 0.11 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr1_-_1356719 | 0.11 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr11_-_2906979 | 0.10 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr19_-_39826639 | 0.10 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr10_+_81466084 | 0.10 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr7_+_6071007 | 0.10 |
ENST00000409061.1
|
ANKRD61
|
ankyrin repeat domain 61 |
| chr22_+_23243156 | 0.09 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr16_+_22524379 | 0.09 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr8_-_131028869 | 0.09 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr4_+_164415855 | 0.09 |
ENST00000508268.1
|
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr16_-_23724518 | 0.09 |
ENST00000457008.2
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr6_-_110500826 | 0.09 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr17_+_30348024 | 0.09 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr1_+_201592013 | 0.09 |
ENST00000593583.1
|
AC096677.1
|
Uncharacterized protein ENSP00000471857 |
| chr22_+_23248512 | 0.09 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr12_+_93861264 | 0.09 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr10_+_91461337 | 0.08 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr10_+_180643 | 0.08 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr22_+_22676808 | 0.08 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_+_196521845 | 0.08 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_-_197686847 | 0.08 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr10_+_5726764 | 0.08 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr4_+_103790462 | 0.08 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr19_+_7587555 | 0.08 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr4_-_103790026 | 0.08 |
ENST00000338145.3
ENST00000357194.6 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_53400813 | 0.08 |
ENST00000595635.1
ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320
|
zinc finger protein 320 |
| chr14_-_105531759 | 0.08 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr11_-_47470703 | 0.08 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr4_+_128982416 | 0.08 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr10_+_48359344 | 0.08 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr16_-_71496112 | 0.08 |
ENST00000393539.2
ENST00000417828.1 ENST00000565718.1 ENST00000497160.1 ENST00000428724.2 |
ZNF23
|
zinc finger protein 23 |
| chr12_+_64845864 | 0.08 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr3_-_182833863 | 0.08 |
ENST00000492597.1
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr8_+_67687413 | 0.07 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr11_-_119293903 | 0.07 |
ENST00000580275.1
|
THY1
|
Thy-1 cell surface antigen |
| chr11_-_64889252 | 0.07 |
ENST00000525297.1
ENST00000529259.1 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr20_-_23669590 | 0.07 |
ENST00000217423.3
|
CST4
|
cystatin S |
| chrX_-_55187531 | 0.07 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr1_+_161494036 | 0.07 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr6_+_109416684 | 0.07 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr19_-_53606604 | 0.07 |
ENST00000599056.1
ENST00000599247.1 ENST00000355147.5 ENST00000429604.1 ENST00000418871.1 ENST00000599637.1 |
ZNF160
|
zinc finger protein 160 |
| chr19_-_45748628 | 0.07 |
ENST00000590022.1
|
AC006126.4
|
AC006126.4 |
| chr10_+_135340859 | 0.07 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr12_-_76742183 | 0.07 |
ENST00000393262.3
|
BBS10
|
Bardet-Biedl syndrome 10 |
| chr8_+_142264664 | 0.07 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr16_+_2880157 | 0.07 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr6_-_74161977 | 0.07 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr4_+_24661479 | 0.07 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr10_+_1034338 | 0.07 |
ENST00000360803.4
ENST00000538293.1 |
GTPBP4
|
GTP binding protein 4 |
| chr10_+_131934643 | 0.07 |
ENST00000331244.5
ENST00000368644.1 |
GLRX3
|
glutaredoxin 3 |
| chr12_+_82752283 | 0.07 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chrX_-_39923656 | 0.07 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr2_+_71357744 | 0.07 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr21_-_44898015 | 0.07 |
ENST00000332440.3
|
LINC00313
|
long intergenic non-protein coding RNA 313 |
| chr5_+_86415919 | 0.07 |
ENST00000515750.1
|
RP11-72L22.1
|
RP11-72L22.1 |
| chr5_+_178450753 | 0.07 |
ENST00000444149.2
ENST00000519896.1 ENST00000522442.1 |
ZNF879
|
zinc finger protein 879 |
| chr12_+_64846129 | 0.07 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr4_+_164415594 | 0.07 |
ENST00000509657.1
ENST00000358572.5 |
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr11_-_95523500 | 0.07 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr19_-_29218538 | 0.07 |
ENST00000592347.1
ENST00000586528.1 |
AC005307.3
|
AC005307.3 |
| chr6_+_2988199 | 0.07 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chr10_-_31320840 | 0.07 |
ENST00000375311.1
|
ZNF438
|
zinc finger protein 438 |
| chr16_+_85204882 | 0.07 |
ENST00000574293.1
|
CTC-786C10.1
|
CTC-786C10.1 |
| chr11_-_93271058 | 0.06 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr17_+_48046671 | 0.06 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr3_+_16926441 | 0.06 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr14_+_52118694 | 0.06 |
ENST00000554778.1
|
FRMD6
|
FERM domain containing 6 |
| chr4_-_7436671 | 0.06 |
ENST00000319098.4
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene) |
| chrX_+_133371077 | 0.06 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr19_-_13068012 | 0.06 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr20_+_24449821 | 0.06 |
ENST00000376862.3
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr14_-_107219365 | 0.06 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr15_-_64338521 | 0.06 |
ENST00000457488.1
ENST00000558069.1 |
DAPK2
|
death-associated protein kinase 2 |
| chr2_-_46844242 | 0.06 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr14_-_75530693 | 0.06 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr2_-_121624973 | 0.06 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chrX_+_118892545 | 0.06 |
ENST00000343905.3
|
SOWAHD
|
sosondowah ankyrin repeat domain family member D |
| chr15_+_33010175 | 0.06 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr4_-_187644930 | 0.06 |
ENST00000441802.2
|
FAT1
|
FAT atypical cadherin 1 |
| chr2_-_87248975 | 0.06 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr1_+_7844312 | 0.06 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr4_-_682769 | 0.06 |
ENST00000507165.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr16_+_1306060 | 0.06 |
ENST00000397534.2
|
TPSD1
|
tryptase delta 1 |
| chr16_+_226658 | 0.06 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr12_+_49760639 | 0.06 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr19_-_53193731 | 0.06 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr3_+_49977440 | 0.06 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr17_-_10633291 | 0.06 |
ENST00000578345.1
ENST00000455996.2 |
TMEM220
|
transmembrane protein 220 |
| chr6_+_168434678 | 0.06 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr18_+_21452804 | 0.06 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr12_+_10366223 | 0.06 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr16_-_3355645 | 0.06 |
ENST00000396862.1
ENST00000573608.1 |
TIGD7
|
tigger transposable element derived 7 |
| chr5_-_180018540 | 0.06 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr10_-_104001231 | 0.06 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr10_+_112327425 | 0.06 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr15_-_77712429 | 0.06 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr8_-_144442136 | 0.06 |
ENST00000519148.1
|
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr3_-_57113281 | 0.06 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr9_+_128510454 | 0.06 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr2_+_204103733 | 0.06 |
ENST00000443941.1
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr4_-_668108 | 0.06 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr18_+_21269404 | 0.06 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr19_-_10450287 | 0.06 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr7_-_120497178 | 0.06 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr11_-_74204692 | 0.06 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr8_-_119634141 | 0.06 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr8_-_97273807 | 0.06 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr17_+_63096903 | 0.06 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr6_+_12008986 | 0.06 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr8_-_131028660 | 0.06 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr7_+_155090271 | 0.06 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr4_+_39640754 | 0.06 |
ENST00000529094.1
ENST00000533736.1 |
RP11-539G18.2
|
RP11-539G18.2 |
| chr5_+_148651409 | 0.06 |
ENST00000296721.4
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr9_-_107754034 | 0.06 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr6_-_166796461 | 0.06 |
ENST00000360961.6
ENST00000341756.6 |
MPC1
|
mitochondrial pyruvate carrier 1 |
| chr19_+_840963 | 0.06 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chr1_-_98510843 | 0.06 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr13_-_22033392 | 0.06 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr1_+_26503894 | 0.06 |
ENST00000361530.6
ENST00000374253.5 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chrY_-_284779 | 0.06 |
ENSTR0000381625.4
|
PPP2R3B
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr5_-_68665296 | 0.05 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr16_-_66835480 | 0.05 |
ENST00000559050.1
ENST00000558713.2 ENST00000433154.1 ENST00000432602.1 ENST00000433574.1 ENST00000415744.1 |
CCDC79
|
coiled-coil domain containing 79 |
| chr4_+_128982430 | 0.05 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr3_-_138553779 | 0.05 |
ENST00000461451.1
|
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr6_-_52109335 | 0.05 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr11_+_8040739 | 0.05 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr3_-_49893907 | 0.05 |
ENST00000482582.1
|
TRAIP
|
TRAF interacting protein |
| chr19_+_11350278 | 0.05 |
ENST00000252453.8
|
C19orf80
|
chromosome 19 open reading frame 80 |
| chr8_+_110551925 | 0.05 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr9_+_35792151 | 0.05 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr12_+_112451222 | 0.05 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr5_-_40798473 | 0.05 |
ENST00000354209.3
|
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr15_+_44084503 | 0.05 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr11_+_121163466 | 0.05 |
ENST00000527762.1
ENST00000534230.1 ENST00000392789.2 |
SC5D
|
sterol-C5-desaturase |
| chr5_+_68665608 | 0.05 |
ENST00000509734.1
ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17
|
RAD17 homolog (S. pombe) |
| chr16_-_82203780 | 0.05 |
ENST00000563504.1
ENST00000569021.1 ENST00000258169.4 |
MPHOSPH6
|
M-phase phosphoprotein 6 |
| chr19_-_8567505 | 0.05 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr9_-_21305312 | 0.05 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr19_-_49956728 | 0.05 |
ENST00000601825.1
ENST00000596049.1 ENST00000599366.1 ENST00000597415.1 |
PIH1D1
|
PIH1 domain containing 1 |
| chr19_+_37960240 | 0.05 |
ENST00000388801.3
|
ZNF570
|
zinc finger protein 570 |
| chr11_-_108338218 | 0.05 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr7_+_149416439 | 0.05 |
ENST00000497895.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr19_+_53970970 | 0.05 |
ENST00000468450.1
ENST00000396403.4 ENST00000490956.1 ENST00000396421.4 |
ZNF813
|
zinc finger protein 813 |
| chr19_+_46236509 | 0.05 |
ENST00000457052.2
|
AC074212.3
|
Uncharacterized protein |
| chr6_-_30709980 | 0.05 |
ENST00000416018.1
ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1
|
flotillin 1 |
| chr4_+_103790120 | 0.05 |
ENST00000273986.4
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr4_-_6694189 | 0.05 |
ENST00000596858.1
|
AC093323.1
|
Uncharacterized protein |
| chr17_-_6554747 | 0.05 |
ENST00000574128.1
|
MED31
|
mediator complex subunit 31 |
| chr19_+_45281118 | 0.05 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr12_+_82752647 | 0.05 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chr19_-_12662314 | 0.05 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr8_-_124428569 | 0.05 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr7_+_64838712 | 0.05 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr7_+_23145347 | 0.05 |
ENST00000322231.7
|
KLHL7
|
kelch-like family member 7 |
| chr16_-_1429010 | 0.05 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr6_+_10694900 | 0.05 |
ENST00000379568.3
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr3_-_47324242 | 0.05 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr17_-_76837499 | 0.05 |
ENST00000592275.1
|
USP36
|
ubiquitin specific peptidase 36 |
| chr19_-_40732594 | 0.05 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr6_-_10694766 | 0.05 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chrX_-_55187588 | 0.05 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr15_-_68497657 | 0.05 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr1_+_47603109 | 0.05 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr12_-_104531785 | 0.05 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr2_-_201753717 | 0.05 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr10_+_60145155 | 0.05 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr22_+_21321531 | 0.05 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr2_-_44223138 | 0.05 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr2_+_121493717 | 0.05 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr10_-_31320860 | 0.05 |
ENST00000436087.2
ENST00000442986.1 ENST00000413025.1 ENST00000452305.1 |
ZNF438
|
zinc finger protein 438 |
| chr6_+_71276596 | 0.05 |
ENST00000370474.3
|
C6orf57
|
chromosome 6 open reading frame 57 |
| chr6_-_31782813 | 0.05 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr19_+_17326191 | 0.05 |
ENST00000595101.1
ENST00000596136.1 ENST00000379776.4 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr12_+_122356488 | 0.05 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr19_-_52511334 | 0.05 |
ENST00000602063.1
ENST00000597747.1 ENST00000594083.1 ENST00000593650.1 ENST00000599631.1 ENST00000598071.1 ENST00000601178.1 ENST00000376716.5 ENST00000391795.3 |
ZNF615
|
zinc finger protein 615 |
| chr2_-_44223089 | 0.05 |
ENST00000447246.1
ENST00000409946.1 ENST00000409659.1 |
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr22_+_18834324 | 0.05 |
ENST00000342005.4
|
AC008132.13
|
Uncharacterized protein |
| chr18_+_39766626 | 0.05 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr2_+_233925038 | 0.05 |
ENST00000451407.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chrX_-_63615297 | 0.05 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr19_+_42381337 | 0.05 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr10_+_75936444 | 0.05 |
ENST00000372734.3
ENST00000541550.1 |
ADK
|
adenosine kinase |
| chr10_+_60144782 | 0.05 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr8_-_101718991 | 0.05 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_+_49977523 | 0.05 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr5_-_90610200 | 0.05 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr4_+_188916918 | 0.05 |
ENST00000509524.1
ENST00000326866.4 |
ZFP42
|
ZFP42 zinc finger protein |
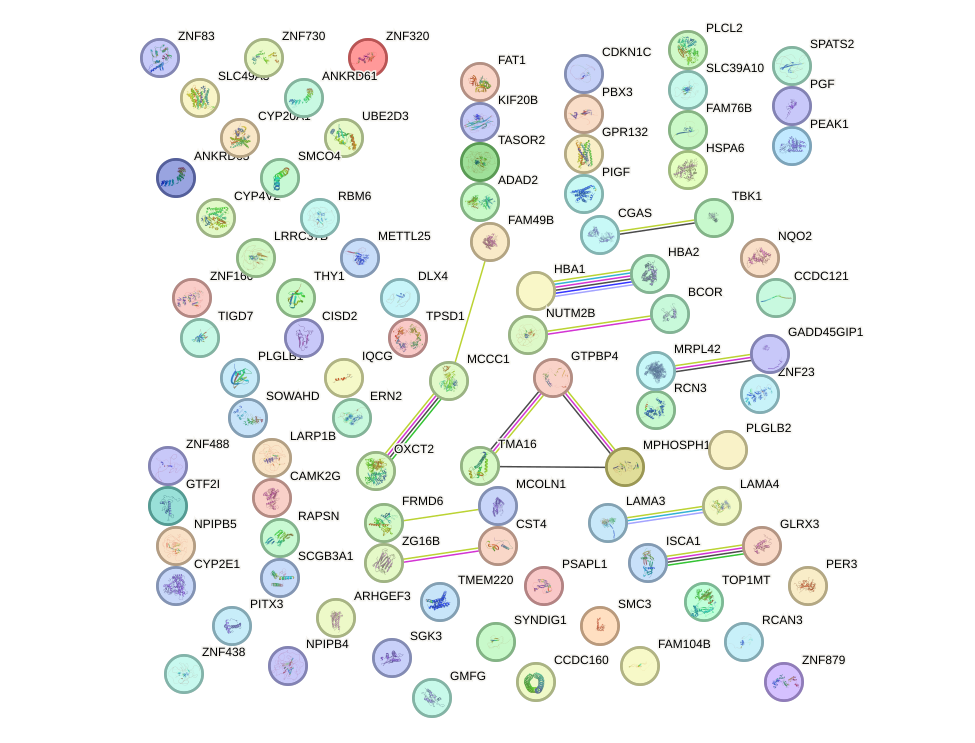
Network of associatons between targets according to the STRING database.
First level regulatory network of MLXIPL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.0 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.0 | 0.1 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.0 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.0 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.0 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.0 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.0 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.0 | GO:0071812 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.3 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.0 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.0 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.1 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.0 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005767 | secondary lysosome(GO:0005767) phagolysosome(GO:0032010) |
| 0.0 | 0.0 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.0 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.0 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.0 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.0 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.0 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |