Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
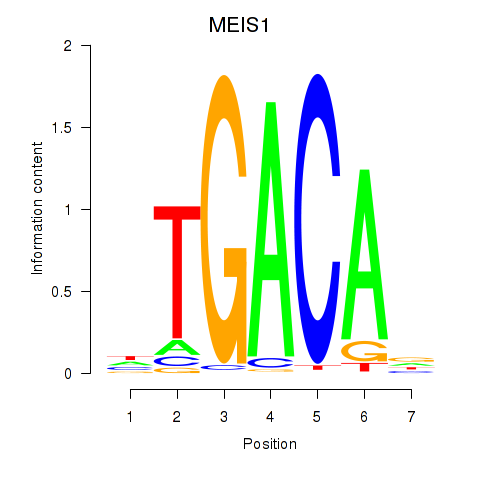
Results for MEIS1
Z-value: 0.32
Transcription factors associated with MEIS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS1
|
ENSG00000143995.15 | Meis homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEIS1 | hg19_v2_chr2_+_66662690_66662711 | 0.85 | 1.5e-01 | Click! |
Activity profile of MEIS1 motif
Sorted Z-values of MEIS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_120365866 | 0.36 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr6_+_53794780 | 0.22 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr1_+_85527987 | 0.20 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr8_-_124286735 | 0.15 |
ENST00000395571.3
|
ZHX1
|
zinc fingers and homeoboxes 1 |
| chr5_-_138780159 | 0.14 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr13_-_44735393 | 0.14 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr8_+_32579271 | 0.13 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr10_+_114709999 | 0.11 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_92053042 | 0.11 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chrX_-_83757399 | 0.11 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr12_-_7281469 | 0.11 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr4_-_83934078 | 0.11 |
ENST00000505397.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr4_+_14113592 | 0.11 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr1_-_247171347 | 0.11 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chrX_+_102024075 | 0.10 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr2_-_202298268 | 0.10 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr19_+_42773371 | 0.10 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr22_-_36013368 | 0.10 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr8_+_81397846 | 0.10 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr14_-_71107921 | 0.10 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr16_-_3350614 | 0.10 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr4_-_83933999 | 0.10 |
ENST00000510557.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr15_+_65337708 | 0.09 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr9_-_20382446 | 0.09 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr4_+_130014836 | 0.09 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr12_+_7055631 | 0.09 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_+_122110691 | 0.09 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr12_-_75784669 | 0.09 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr5_-_65018834 | 0.09 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr17_+_8316442 | 0.09 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr6_-_134861089 | 0.09 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr8_+_32579321 | 0.09 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr17_+_73642315 | 0.09 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chrX_-_70331298 | 0.09 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr7_+_26438187 | 0.08 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr4_+_76871883 | 0.08 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr11_+_10772847 | 0.08 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr1_-_100598444 | 0.08 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr6_-_52109335 | 0.08 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr6_+_144665237 | 0.08 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr12_+_498500 | 0.08 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr5_+_32712363 | 0.08 |
ENST00000507141.1
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr16_+_2588012 | 0.08 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr11_+_35198243 | 0.08 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr7_-_27169801 | 0.08 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr3_+_16926441 | 0.08 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr12_+_95611536 | 0.08 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_+_95612006 | 0.08 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_42927251 | 0.08 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr5_+_60240943 | 0.08 |
ENST00000296597.5
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chrX_-_85302531 | 0.08 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr21_-_43916296 | 0.07 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr10_+_114710516 | 0.07 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_37311588 | 0.07 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr2_+_149402989 | 0.07 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr19_-_47164386 | 0.07 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr1_-_154178803 | 0.07 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr18_+_44526786 | 0.07 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr21_-_43916433 | 0.07 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr14_-_39639523 | 0.07 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr7_+_110731062 | 0.07 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr2_-_24299308 | 0.07 |
ENST00000233468.4
|
SF3B14
|
Pre-mRNA branch site protein p14 |
| chr2_+_24714729 | 0.07 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr2_-_119605253 | 0.07 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr6_-_72129806 | 0.07 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr18_-_2982869 | 0.07 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr8_+_26149274 | 0.07 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr1_-_43282945 | 0.07 |
ENST00000537227.1
|
CCDC23
|
coiled-coil domain containing 23 |
| chr9_-_6015607 | 0.07 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr1_+_171810606 | 0.07 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr9_-_16705069 | 0.06 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr1_+_28099700 | 0.06 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr10_+_86004802 | 0.06 |
ENST00000359452.4
ENST00000358110.5 ENST00000372092.3 |
RGR
|
retinal G protein coupled receptor |
| chr1_-_89357179 | 0.06 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr17_+_44668387 | 0.06 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr8_+_97597148 | 0.06 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chrX_+_149867681 | 0.06 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr8_+_38239882 | 0.06 |
ENST00000607047.1
|
RP11-350N15.5
|
RP11-350N15.5 |
| chr17_-_74528128 | 0.06 |
ENST00000590175.1
|
CYGB
|
cytoglobin |
| chr17_-_77925806 | 0.06 |
ENST00000574241.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr17_-_1029866 | 0.06 |
ENST00000570525.1
|
ABR
|
active BCR-related |
| chr5_+_134368954 | 0.06 |
ENST00000432382.3
|
CTC-349C3.1
|
chromosome 5 open reading frame 66 |
| chr1_-_198906528 | 0.06 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr1_+_154909803 | 0.06 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr22_+_22676808 | 0.06 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr4_-_102268708 | 0.06 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr5_-_102455801 | 0.06 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr16_+_88636875 | 0.06 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr12_-_11002063 | 0.06 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr4_+_77941685 | 0.06 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr11_-_108338218 | 0.06 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr3_+_63897605 | 0.06 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr5_-_168006324 | 0.06 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr20_-_18774614 | 0.06 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr17_+_68165657 | 0.06 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr16_-_1429010 | 0.06 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr12_+_498545 | 0.06 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr11_-_85780853 | 0.06 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr13_+_95364963 | 0.06 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr5_+_96079240 | 0.06 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr16_-_54963026 | 0.06 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr16_-_20566616 | 0.06 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr1_-_26231589 | 0.06 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr3_+_141103634 | 0.06 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_32579341 | 0.06 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr5_+_79783788 | 0.06 |
ENST00000282226.4
|
FAM151B
|
family with sequence similarity 151, member B |
| chr13_-_52027134 | 0.06 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr2_-_42180940 | 0.06 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr3_-_24536222 | 0.06 |
ENST00000415021.1
ENST00000447875.1 |
THRB
|
thyroid hormone receptor, beta |
| chr19_-_4535233 | 0.06 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr20_-_32580924 | 0.06 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr11_-_72852320 | 0.05 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr1_-_109584716 | 0.05 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr13_-_34250861 | 0.05 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr2_-_208030295 | 0.05 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr19_-_10446449 | 0.05 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr16_+_28996572 | 0.05 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr3_+_107318157 | 0.05 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr8_-_74659693 | 0.05 |
ENST00000518767.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr5_+_60241020 | 0.05 |
ENST00000511107.1
ENST00000502658.1 |
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr11_+_17281900 | 0.05 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr3_+_141121164 | 0.05 |
ENST00000510338.1
ENST00000504673.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_61429728 | 0.05 |
ENST00000529579.1
|
RAB2A
|
RAB2A, member RAS oncogene family |
| chr3_-_196014520 | 0.05 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chrX_+_107068959 | 0.05 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr3_-_194373831 | 0.05 |
ENST00000437613.1
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr18_+_56807096 | 0.05 |
ENST00000588875.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr14_+_45464658 | 0.05 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chr12_-_57030096 | 0.05 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr16_+_53164833 | 0.05 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_76203345 | 0.05 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr13_-_52026730 | 0.05 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr5_+_102455853 | 0.05 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_+_21679220 | 0.05 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr1_+_104068562 | 0.05 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr14_-_24701539 | 0.05 |
ENST00000534348.1
ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr17_-_66287310 | 0.05 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr8_-_99837856 | 0.05 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr17_-_66287257 | 0.05 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr13_+_76378357 | 0.05 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr2_+_208576259 | 0.05 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr17_+_19314505 | 0.05 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr9_-_132597529 | 0.05 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr17_+_62503147 | 0.05 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chr10_-_131762105 | 0.05 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr1_-_150978953 | 0.05 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr22_+_23248512 | 0.05 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr10_-_21186144 | 0.05 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_-_118810688 | 0.05 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr14_-_58893832 | 0.05 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chrX_-_43832711 | 0.05 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr1_-_91317072 | 0.05 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr2_+_201170770 | 0.05 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr21_-_47352477 | 0.05 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr14_+_58894404 | 0.05 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr12_-_51785182 | 0.05 |
ENST00000356317.3
ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr6_+_90272488 | 0.05 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr17_+_26833250 | 0.05 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr15_-_71407806 | 0.05 |
ENST00000566432.1
ENST00000567117.1 |
CT62
|
cancer/testis antigen 62 |
| chr15_-_71407833 | 0.05 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr8_-_42360015 | 0.05 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr15_+_44069276 | 0.05 |
ENST00000381359.1
|
SERF2
|
small EDRK-rich factor 2 |
| chr15_-_34502197 | 0.05 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr14_-_50154921 | 0.05 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr14_+_57735636 | 0.05 |
ENST00000556995.1
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chrX_-_54070388 | 0.05 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr9_+_127615733 | 0.05 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr5_+_49962772 | 0.05 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr9_+_2157655 | 0.05 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_153191706 | 0.05 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr4_+_159593418 | 0.05 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr13_-_54706954 | 0.05 |
ENST00000606706.1
ENST00000607494.1 ENST00000427299.2 ENST00000423442.2 ENST00000451744.1 |
LINC00458
|
long intergenic non-protein coding RNA 458 |
| chr4_-_123542224 | 0.05 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr3_+_9774164 | 0.05 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr7_-_32111009 | 0.05 |
ENST00000396184.3
ENST00000396189.2 ENST00000321453.7 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr1_+_236557569 | 0.05 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr12_+_19358228 | 0.05 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_-_166796461 | 0.05 |
ENST00000360961.6
ENST00000341756.6 |
MPC1
|
mitochondrial pyruvate carrier 1 |
| chrX_+_102965835 | 0.04 |
ENST00000319560.6
|
TMEM31
|
transmembrane protein 31 |
| chr22_+_21213259 | 0.04 |
ENST00000215730.7
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr10_-_119805558 | 0.04 |
ENST00000369199.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr8_+_99076509 | 0.04 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chrX_+_150148976 | 0.04 |
ENST00000419110.1
|
HMGB3
|
high mobility group box 3 |
| chr7_+_77428066 | 0.04 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr2_+_242312264 | 0.04 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr11_-_122929699 | 0.04 |
ENST00000526686.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr1_-_1590418 | 0.04 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr4_-_83931862 | 0.04 |
ENST00000506560.1
ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54
|
lin-54 homolog (C. elegans) |
| chr20_+_52824367 | 0.04 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr4_+_77870960 | 0.04 |
ENST00000505788.1
ENST00000510515.1 ENST00000504637.1 |
SEPT11
|
septin 11 |
| chr1_+_169337412 | 0.04 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr16_+_57702210 | 0.04 |
ENST00000450388.3
|
GPR97
|
G protein-coupled receptor 97 |
| chr12_+_62654155 | 0.04 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr17_-_62503015 | 0.04 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_-_54872059 | 0.04 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr2_-_230787879 | 0.04 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr15_-_70994612 | 0.04 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_-_23053693 | 0.04 |
ENST00000409763.1
ENST00000409923.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr19_+_36266417 | 0.04 |
ENST00000378944.5
ENST00000007510.4 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr15_+_58724184 | 0.04 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.0 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.2 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.0 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0045924 | regulation of female receptivity(GO:0045924) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.0 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.0 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.0 | 0.0 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0097198 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.0 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.0 | GO:0042599 | lamellar body(GO:0042599) alveolar lamellar body(GO:0097208) lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |