Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
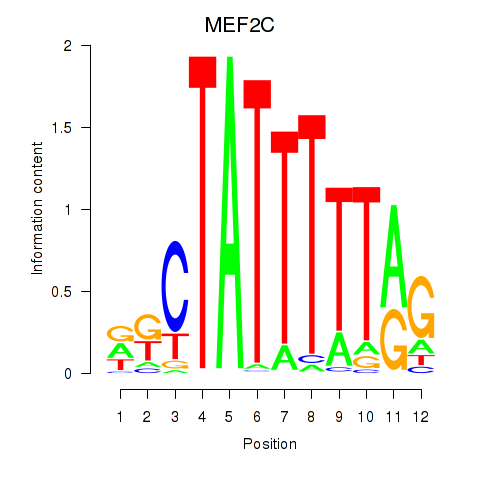
Results for MEF2C
Z-value: 0.56
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2C | hg19_v2_chr5_-_88179017_88179050 | 0.50 | 5.0e-01 | Click! |
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_18923035 | 0.19 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr21_+_35107346 | 0.17 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr8_+_107282368 | 0.16 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr4_-_170192000 | 0.16 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr1_+_198608146 | 0.15 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr10_-_45474237 | 0.15 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr20_-_44519839 | 0.15 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr12_-_111358372 | 0.15 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr15_-_76352069 | 0.14 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr17_+_7184986 | 0.14 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr21_+_17909594 | 0.13 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_+_18725758 | 0.13 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr15_+_63889577 | 0.13 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr10_+_71561630 | 0.13 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr7_+_143013198 | 0.12 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr12_-_49393092 | 0.12 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr3_+_159570722 | 0.11 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr15_+_63889552 | 0.11 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chrX_-_15683147 | 0.11 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr17_-_48785216 | 0.11 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr8_+_101349823 | 0.10 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr17_+_4855053 | 0.10 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr11_+_65266507 | 0.10 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_-_57644952 | 0.10 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr22_+_19706958 | 0.09 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr17_-_34625719 | 0.09 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr2_+_173686303 | 0.09 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr18_+_3252265 | 0.09 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_+_3666335 | 0.09 |
ENST00000250693.1
|
ART1
|
ADP-ribosyltransferase 1 |
| chr15_+_78830023 | 0.09 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr7_+_152456904 | 0.08 |
ENST00000537264.1
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr1_-_59249732 | 0.08 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr7_+_95115210 | 0.08 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr16_+_31225337 | 0.08 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr5_+_96077888 | 0.08 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
| chr2_+_182850743 | 0.08 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr17_-_34524157 | 0.08 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr12_+_54384370 | 0.08 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr9_+_130026756 | 0.08 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr11_-_104480019 | 0.08 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr1_-_226187013 | 0.08 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr22_-_31328881 | 0.08 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr11_+_128563652 | 0.07 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_191592 | 0.07 |
ENST00000328278.3
|
LRRC14B
|
leucine rich repeat containing 14B |
| chr7_+_152456829 | 0.07 |
ENST00000377776.3
ENST00000256001.8 ENST00000397282.2 |
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr16_+_30075783 | 0.07 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chrX_+_135279179 | 0.07 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr6_+_151358048 | 0.07 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr14_+_76776957 | 0.07 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr2_+_145780739 | 0.07 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr16_+_30075595 | 0.07 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_-_62457371 | 0.07 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr8_+_95565947 | 0.07 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr3_+_69812877 | 0.07 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr4_+_37892682 | 0.07 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr10_+_71562180 | 0.07 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr12_+_53846594 | 0.07 |
ENST00000550192.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr10_+_71561649 | 0.06 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr15_+_100106244 | 0.06 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr8_-_73793975 | 0.06 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr10_-_75410771 | 0.06 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr14_-_74462922 | 0.06 |
ENST00000553284.1
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr22_-_36031181 | 0.06 |
ENST00000594060.1
|
AL049747.1
|
AL049747.1 |
| chr5_-_16509101 | 0.06 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr15_+_100106126 | 0.06 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr3_-_192445289 | 0.06 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr10_-_61495760 | 0.06 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr10_-_75415825 | 0.06 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr3_+_141106643 | 0.06 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_-_148778323 | 0.05 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr4_-_88450612 | 0.05 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr9_-_110251836 | 0.05 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr1_-_100643765 | 0.05 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr2_+_109204909 | 0.05 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_+_42651691 | 0.05 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr22_-_51016846 | 0.05 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr12_+_40618764 | 0.05 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr1_+_16010779 | 0.05 |
ENST00000375799.3
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr2_+_103378472 | 0.05 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr10_+_124134201 | 0.05 |
ENST00000368990.3
ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr3_-_176914238 | 0.05 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr22_-_51017084 | 0.05 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr17_+_65040678 | 0.05 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr3_-_39234074 | 0.05 |
ENST00000340369.3
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin-binding repeat containing 1 |
| chr1_+_156096336 | 0.05 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr4_+_39408470 | 0.05 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr3_+_141106458 | 0.05 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_-_17533838 | 0.04 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr9_+_116327326 | 0.04 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chrX_-_142722897 | 0.04 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr1_-_205782304 | 0.04 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr11_-_47399942 | 0.04 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_+_156095951 | 0.04 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr12_-_131200810 | 0.04 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr11_-_47400062 | 0.04 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_+_65663812 | 0.04 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr15_+_80364901 | 0.04 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr8_+_132952112 | 0.04 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr2_-_148779106 | 0.04 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr16_+_30075463 | 0.04 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_-_63782888 | 0.04 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr3_-_4927447 | 0.04 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr11_-_47400078 | 0.04 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr6_+_123038689 | 0.04 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr2_+_233497931 | 0.04 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chrX_+_135278908 | 0.04 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr3_-_108248169 | 0.04 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr8_+_76452097 | 0.04 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr20_+_3767547 | 0.03 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr3_+_193853927 | 0.03 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr14_+_39735411 | 0.03 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr3_-_30936153 | 0.03 |
ENST00000454381.3
ENST00000282538.5 |
GADL1
|
glutamate decarboxylase-like 1 |
| chr5_-_131347306 | 0.03 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr3_+_49057876 | 0.03 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_-_49576198 | 0.03 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr20_+_44035847 | 0.03 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_69924639 | 0.03 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr10_+_88428206 | 0.03 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr2_+_239047337 | 0.03 |
ENST00000409223.1
ENST00000305959.4 |
KLHL30
|
kelch-like family member 30 |
| chr20_+_44657845 | 0.03 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr12_-_122240792 | 0.03 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr8_+_1993173 | 0.03 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr3_+_69928256 | 0.03 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_145525015 | 0.03 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr11_+_7598239 | 0.03 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_+_24018269 | 0.03 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr1_+_41174988 | 0.03 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr3_+_113616317 | 0.03 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr4_-_185303418 | 0.03 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr4_+_110834033 | 0.03 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr2_-_151344172 | 0.03 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr11_-_85780853 | 0.03 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_917466 | 0.03 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr7_-_56160666 | 0.03 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr6_+_161123270 | 0.03 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr5_-_150521192 | 0.03 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr14_+_39736582 | 0.03 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr18_+_12407895 | 0.03 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr1_+_41447609 | 0.03 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chrX_+_37545012 | 0.03 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr12_-_46766577 | 0.03 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr2_-_231825668 | 0.03 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr5_-_137475071 | 0.03 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr9_+_100263912 | 0.03 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr2_-_148778258 | 0.03 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr8_+_1993152 | 0.03 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr8_-_66701319 | 0.03 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chrX_-_119693745 | 0.03 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr1_-_150693305 | 0.03 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr13_-_30880979 | 0.03 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr5_+_138609782 | 0.03 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr9_-_16728161 | 0.03 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr11_+_75110530 | 0.03 |
ENST00000531188.1
ENST00000530164.1 ENST00000422465.2 ENST00000278572.6 ENST00000534440.1 ENST00000527446.1 ENST00000526608.1 ENST00000527273.1 ENST00000524851.1 |
RPS3
|
ribosomal protein S3 |
| chr14_-_65409502 | 0.02 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr18_+_9708162 | 0.02 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr1_-_713985 | 0.02 |
ENST00000428504.1
|
RP11-206L10.2
|
RP11-206L10.2 |
| chr22_+_41487711 | 0.02 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr3_-_38071122 | 0.02 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr1_-_150693318 | 0.02 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chrX_-_131353461 | 0.02 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr10_+_88428370 | 0.02 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr15_-_69113218 | 0.02 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr19_+_37096194 | 0.02 |
ENST00000460670.1
ENST00000292928.2 ENST00000439428.1 |
ZNF382
|
zinc finger protein 382 |
| chr8_-_94029882 | 0.02 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr4_+_160203650 | 0.02 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr5_-_111091948 | 0.02 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr1_+_145524891 | 0.02 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr9_+_127054254 | 0.02 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr3_-_11610255 | 0.02 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr10_+_133753533 | 0.02 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chrX_+_153046456 | 0.02 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr1_-_154164534 | 0.02 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr14_+_24540731 | 0.02 |
ENST00000558859.1
ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6
|
copine VI (neuronal) |
| chr10_+_63661053 | 0.02 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr19_-_51220176 | 0.02 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr5_-_131347583 | 0.02 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr20_+_61147651 | 0.02 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr20_+_36373032 | 0.02 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr11_+_77774897 | 0.02 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr7_-_82073109 | 0.02 |
ENST00000356860.3
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr10_+_118083919 | 0.02 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr12_-_118406028 | 0.02 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr22_+_32340481 | 0.02 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr9_+_127023704 | 0.02 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr4_-_143767428 | 0.02 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_+_22225041 | 0.02 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr21_+_27011584 | 0.02 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr3_+_4535025 | 0.02 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr7_-_78400364 | 0.02 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr15_-_83474806 | 0.02 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr7_-_30739661 | 0.02 |
ENST00000445981.1
ENST00000348438.4 |
CRHR2
|
corticotropin releasing hormone receptor 2 |
| chr1_+_203097407 | 0.02 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr6_-_161085291 | 0.01 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr18_-_3219847 | 0.01 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr17_-_27467418 | 0.01 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr17_-_1389419 | 0.01 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr15_-_93353028 | 0.01 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr17_+_7461580 | 0.01 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr21_+_35552978 | 0.01 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr6_-_154751629 | 0.01 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044027 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.0 | GO:1903216 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |