Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
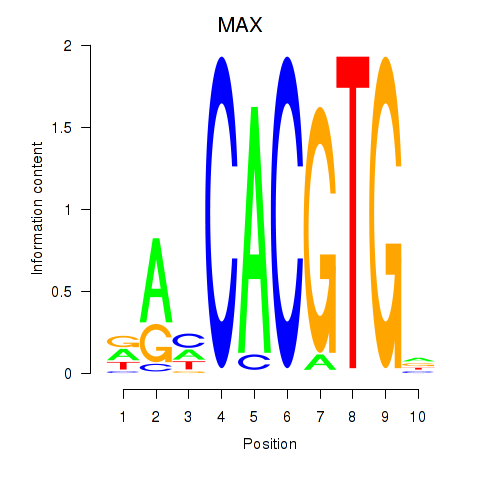
Results for MAX_TFEB
Z-value: 0.75
Transcription factors associated with MAX_TFEB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAX
|
ENSG00000125952.14 | MYC associated factor X |
|
TFEB
|
ENSG00000112561.13 | transcription factor EB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFEB | hg19_v2_chr6_-_41673552_41673678 | 0.90 | 9.9e-02 | Click! |
| MAX | hg19_v2_chr14_-_65569057_65569119 | -0.88 | 1.2e-01 | Click! |
Activity profile of MAX_TFEB motif
Sorted Z-values of MAX_TFEB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_33385854 | 1.34 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_-_7137857 | 1.23 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr16_+_2570340 | 1.19 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr1_+_44440575 | 1.05 |
ENST00000532642.1
ENST00000236067.4 ENST00000471859.2 |
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr17_-_7137582 | 0.94 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr19_-_10764509 | 0.93 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr12_-_58146048 | 0.78 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr1_-_11865982 | 0.74 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr19_-_36545649 | 0.71 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr8_-_103876340 | 0.63 |
ENST00000518353.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr16_-_5083917 | 0.63 |
ENST00000312251.3
ENST00000381955.3 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr14_-_20922960 | 0.62 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr16_-_5083589 | 0.61 |
ENST00000563578.1
ENST00000562346.2 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr10_-_76995769 | 0.60 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr6_-_44225231 | 0.59 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr2_+_220042933 | 0.59 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr12_+_54332535 | 0.58 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chr10_-_76995675 | 0.56 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr7_+_150759634 | 0.55 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr3_+_19988736 | 0.55 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr9_-_131709858 | 0.53 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr2_-_220042825 | 0.52 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr16_-_1525016 | 0.49 |
ENST00000262318.8
ENST00000448525.1 |
CLCN7
|
chloride channel, voltage-sensitive 7 |
| chr6_-_43197189 | 0.49 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr16_-_28503327 | 0.48 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr14_-_20929624 | 0.48 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr17_+_62223320 | 0.46 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr3_-_196987309 | 0.45 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_+_99258625 | 0.45 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr12_-_58146128 | 0.45 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr4_-_2935674 | 0.44 |
ENST00000514800.1
|
MFSD10
|
major facilitator superfamily domain containing 10 |
| chr20_-_62493217 | 0.44 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr5_-_176730733 | 0.43 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chr20_-_2821271 | 0.42 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr19_-_47734448 | 0.41 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr2_-_47572105 | 0.40 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr12_-_49463753 | 0.40 |
ENST00000301068.6
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr16_+_2570431 | 0.39 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr19_+_1275917 | 0.39 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr19_-_5680891 | 0.38 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr1_+_11796177 | 0.38 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr16_-_28503080 | 0.37 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr12_-_121342170 | 0.37 |
ENST00000353487.2
|
SPPL3
|
signal peptide peptidase like 3 |
| chr11_+_67159416 | 0.37 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr11_+_57509020 | 0.36 |
ENST00000388857.4
ENST00000528798.1 |
C11orf31
|
chromosome 11 open reading frame 31 |
| chr1_+_92414952 | 0.36 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr11_+_65627974 | 0.36 |
ENST00000525768.1
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr1_+_11796126 | 0.36 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr4_-_100871506 | 0.36 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr17_+_6915798 | 0.35 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr13_+_113951607 | 0.35 |
ENST00000397181.3
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr17_+_6915730 | 0.35 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr1_-_154193091 | 0.34 |
ENST00000362076.4
ENST00000350592.3 ENST00000368516.1 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr19_+_49458107 | 0.34 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr19_-_40324767 | 0.33 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_-_11545920 | 0.33 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr5_+_149865838 | 0.33 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr12_+_56110315 | 0.33 |
ENST00000548556.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr11_-_71814422 | 0.33 |
ENST00000278671.5
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr16_+_28985542 | 0.33 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr3_-_178789220 | 0.33 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr19_-_41256207 | 0.32 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr6_-_33385655 | 0.32 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr19_+_41305330 | 0.31 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr2_-_47572207 | 0.31 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr19_+_14544099 | 0.31 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr3_+_183967409 | 0.31 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr15_-_72668805 | 0.31 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr8_-_144679264 | 0.31 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr16_+_28986085 | 0.31 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr5_+_150827143 | 0.31 |
ENST00000243389.3
ENST00000517945.1 ENST00000521925.1 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr1_-_155232221 | 0.31 |
ENST00000355379.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr13_+_113951532 | 0.30 |
ENST00000332556.4
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr19_-_15236470 | 0.30 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr17_-_38256973 | 0.29 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr1_+_11333546 | 0.29 |
ENST00000376804.2
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr16_+_57220049 | 0.29 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr17_+_1627834 | 0.28 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr11_+_57508825 | 0.28 |
ENST00000534355.1
|
C11orf31
|
chromosome 11 open reading frame 31 |
| chr17_+_72428266 | 0.28 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr16_-_28503357 | 0.28 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chrX_+_153627231 | 0.28 |
ENST00000406022.2
|
RPL10
|
ribosomal protein L10 |
| chr18_+_20715416 | 0.28 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr17_+_42148097 | 0.28 |
ENST00000269097.4
|
G6PC3
|
glucose 6 phosphatase, catalytic, 3 |
| chr17_+_46970178 | 0.28 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr16_-_88923285 | 0.28 |
ENST00000542788.1
ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr3_-_4508925 | 0.28 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr19_-_41903161 | 0.27 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr3_+_51428704 | 0.27 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chrY_-_15591818 | 0.27 |
ENST00000382893.1
|
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr11_-_1785139 | 0.27 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr7_+_99699280 | 0.27 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr7_+_4815238 | 0.26 |
ENST00000348624.4
ENST00000401897.1 |
AP5Z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chr12_-_108154705 | 0.26 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr19_-_15236562 | 0.26 |
ENST00000263383.3
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr5_-_154230130 | 0.25 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chrX_-_100662881 | 0.25 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr19_+_7587491 | 0.25 |
ENST00000264079.6
|
MCOLN1
|
mucolipin 1 |
| chr3_-_49395892 | 0.25 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr7_+_99699179 | 0.25 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr17_+_6915902 | 0.25 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr6_-_33385870 | 0.25 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_-_18266818 | 0.25 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr11_+_62538775 | 0.25 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr15_+_75628232 | 0.25 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr12_-_53625958 | 0.24 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr14_-_54420133 | 0.24 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr22_-_39268192 | 0.24 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr11_+_61560348 | 0.24 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr9_+_131709966 | 0.24 |
ENST00000372577.2
|
NUP188
|
nucleoporin 188kDa |
| chr17_+_74732889 | 0.24 |
ENST00000591864.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr15_+_89182156 | 0.24 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr20_-_2821756 | 0.24 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr6_-_31763276 | 0.23 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr2_-_198364552 | 0.23 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr19_-_49016847 | 0.23 |
ENST00000598924.1
|
CTC-273B12.10
|
CTC-273B12.10 |
| chrX_+_23685653 | 0.23 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr22_-_42343117 | 0.23 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr19_-_40854281 | 0.23 |
ENST00000392035.2
|
C19orf47
|
chromosome 19 open reading frame 47 |
| chr19_+_6464243 | 0.23 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr6_-_33385823 | 0.23 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr19_+_39903185 | 0.23 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr16_+_5121814 | 0.22 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr12_+_6644443 | 0.22 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr19_+_50016411 | 0.22 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_-_10426663 | 0.22 |
ENST00000541276.1
ENST00000393708.3 ENST00000494368.1 |
FDX1L
|
ferredoxin 1-like |
| chr7_+_100271355 | 0.22 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr11_+_67776012 | 0.22 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr22_+_31002779 | 0.22 |
ENST00000215838.3
|
TCN2
|
transcobalamin II |
| chr1_-_212873267 | 0.22 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr19_-_4670345 | 0.22 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr9_-_34637806 | 0.21 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr7_+_100271446 | 0.21 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr22_+_31003133 | 0.21 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr10_+_99344071 | 0.21 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr19_+_40854363 | 0.21 |
ENST00000599685.1
ENST00000392032.2 |
PLD3
|
phospholipase D family, member 3 |
| chr15_+_75628394 | 0.21 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr19_+_40854559 | 0.21 |
ENST00000598962.1
ENST00000409419.1 ENST00000409587.1 ENST00000602131.1 ENST00000409735.4 ENST00000600948.1 ENST00000356508.5 ENST00000596682.1 ENST00000594908.1 |
PLD3
|
phospholipase D family, member 3 |
| chr16_-_1401799 | 0.21 |
ENST00000007390.2
|
TSR3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr1_-_153940097 | 0.21 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr3_+_72201910 | 0.21 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr11_-_36310958 | 0.21 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr7_+_2559399 | 0.21 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_10093188 | 0.21 |
ENST00000377153.1
|
UBE4B
|
ubiquitination factor E4B |
| chr15_+_75628419 | 0.21 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr6_+_42018614 | 0.21 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr16_+_5083950 | 0.21 |
ENST00000588623.1
|
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr2_-_85829780 | 0.21 |
ENST00000334462.5
|
TMEM150A
|
transmembrane protein 150A |
| chr9_-_140009130 | 0.21 |
ENST00000497375.1
ENST00000371579.2 |
DPP7
|
dipeptidyl-peptidase 7 |
| chr22_+_35776828 | 0.20 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr7_+_99070527 | 0.20 |
ENST00000379724.3
|
ZNF789
|
zinc finger protein 789 |
| chr22_-_20850070 | 0.20 |
ENST00000440659.2
ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22
|
kelch-like family member 22 |
| chr6_-_31612808 | 0.20 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr1_-_154193009 | 0.20 |
ENST00000368518.1
ENST00000368519.1 ENST00000368521.5 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr17_-_37886752 | 0.20 |
ENST00000577810.1
|
MIEN1
|
migration and invasion enhancer 1 |
| chr22_+_21996549 | 0.20 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr1_-_111991850 | 0.20 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr19_-_12833164 | 0.20 |
ENST00000356861.5
|
TNPO2
|
transportin 2 |
| chr19_-_51307894 | 0.20 |
ENST00000597705.1
ENST00000391812.1 |
C19orf48
|
chromosome 19 open reading frame 48 |
| chr17_-_4607335 | 0.20 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr19_-_5720248 | 0.20 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr17_-_79520135 | 0.20 |
ENST00000541246.1
ENST00000544302.1 |
C17orf70
|
chromosome 17 open reading frame 70 |
| chr3_-_49395705 | 0.20 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr1_-_207119738 | 0.19 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr7_-_44530479 | 0.19 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr17_+_46018872 | 0.19 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr3_+_50654821 | 0.19 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr19_-_47290535 | 0.19 |
ENST00000412532.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr11_-_66725837 | 0.19 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr16_-_67190152 | 0.19 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr16_+_28986134 | 0.19 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr19_+_45681997 | 0.19 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr20_-_3748363 | 0.19 |
ENST00000217195.8
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr17_+_46970134 | 0.19 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr7_+_148959262 | 0.19 |
ENST00000434415.1
|
ZNF783
|
zinc finger family member 783 |
| chr7_+_150065879 | 0.19 |
ENST00000397281.2
ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr10_-_99258135 | 0.19 |
ENST00000327238.10
ENST00000327277.7 ENST00000355839.6 ENST00000437002.1 ENST00000422685.1 |
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
| chr15_-_72668185 | 0.19 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr10_-_126849626 | 0.19 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr9_+_133978190 | 0.19 |
ENST00000372312.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr12_+_113623325 | 0.18 |
ENST00000549621.1
ENST00000548278.1 ENST00000552495.1 |
C12orf52
|
RBPJ interacting and tubulin associated 1 |
| chr19_-_45909585 | 0.18 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr17_+_72428218 | 0.18 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_-_49967292 | 0.18 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr22_-_20850128 | 0.18 |
ENST00000328879.4
|
KLHL22
|
kelch-like family member 22 |
| chr2_-_85829811 | 0.18 |
ENST00000306353.3
|
TMEM150A
|
transmembrane protein 150A |
| chr3_-_49761337 | 0.18 |
ENST00000535833.1
ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3
GMPPB
|
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr19_+_36545781 | 0.18 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr11_-_71814276 | 0.18 |
ENST00000538404.1
ENST00000535107.1 ENST00000545249.1 |
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr5_+_149865377 | 0.18 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr19_-_45681482 | 0.18 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr12_+_51633061 | 0.18 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr3_-_49055991 | 0.18 |
ENST00000441576.2
ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr19_+_41256764 | 0.18 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr7_+_99746514 | 0.18 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr22_-_42765174 | 0.18 |
ENST00000432473.1
ENST00000412060.1 ENST00000424852.1 |
Z83851.1
|
Z83851.1 |
| chr8_+_103876528 | 0.18 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr4_+_76439649 | 0.18 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr9_-_19127474 | 0.18 |
ENST00000380465.3
ENST00000380464.3 ENST00000411567.1 ENST00000276914.2 |
PLIN2
|
perilipin 2 |
| chr6_-_33385902 | 0.18 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAX_TFEB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 1.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 1.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 0.6 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.2 | 1.2 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 0.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.5 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.2 | 0.6 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 0.8 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 0.5 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.4 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.3 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.1 | 0.3 | GO:1902512 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.4 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.3 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.4 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.8 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 1.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.5 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.2 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.4 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.2 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.2 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.2 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 1.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.4 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.5 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0006818 | hydrogen transport(GO:0006818) proton transport(GO:0015992) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:2000254 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) regulation of male germ cell proliferation(GO:2000254) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.0 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.0 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.0 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.0 | GO:0018013 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 2.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 1.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.2 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 0.5 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.4 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.7 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.4 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.6 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.9 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.2 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.1 | 2.0 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.5 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.2 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.5 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 1.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 1.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 2.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.6 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.0 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |