Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
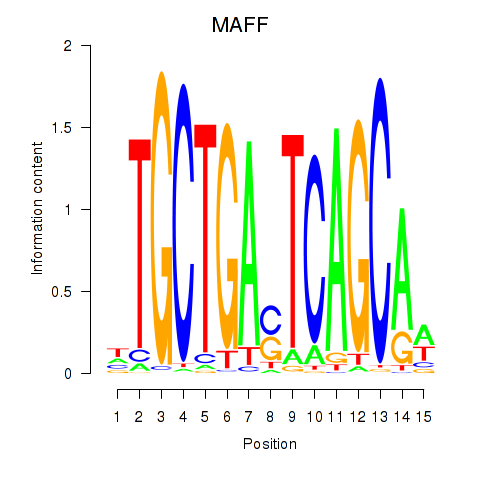
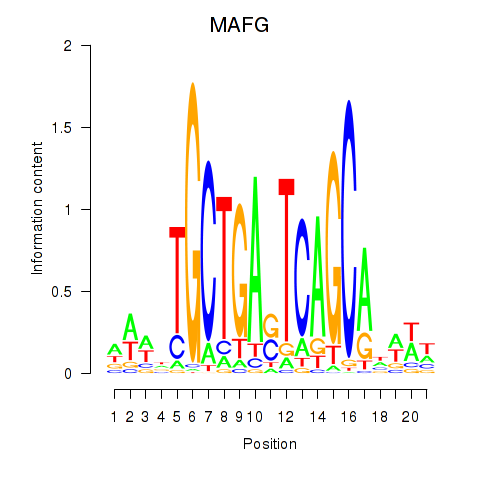
Results for MAFF_MAFG
Z-value: 0.92
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.7 | MAF bZIP transcription factor F |
|
MAFG
|
ENSG00000197063.6 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFF | hg19_v2_chr22_+_38609538_38609547 | -0.81 | 1.9e-01 | Click! |
| MAFG | hg19_v2_chr17_-_79881408_79881423 | 0.44 | 5.6e-01 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_142720267 | 0.78 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr12_-_14924056 | 0.62 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chr1_+_85527987 | 0.59 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr16_-_25122785 | 0.56 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr15_+_23810903 | 0.44 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chrX_+_140096761 | 0.42 |
ENST00000370530.1
|
SPANXB1
|
SPANX family, member B1 |
| chr7_+_5465382 | 0.40 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr14_-_75530693 | 0.40 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr17_-_14140166 | 0.40 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr17_-_79881408 | 0.38 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr14_+_94577074 | 0.38 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr22_+_20905422 | 0.35 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr16_-_87970122 | 0.35 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr6_+_53794780 | 0.33 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr16_-_11723066 | 0.32 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr13_-_44453826 | 0.32 |
ENST00000444614.3
|
CCDC122
|
coiled-coil domain containing 122 |
| chr7_-_86849836 | 0.31 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr16_+_66442411 | 0.31 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr5_-_176449586 | 0.31 |
ENST00000509236.1
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chrX_-_14891150 | 0.29 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr22_+_22556057 | 0.28 |
ENST00000390286.2
|
IGLV11-55
|
immunoglobulin lambda variable 11-55 (non-functional) |
| chr6_+_31638156 | 0.28 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr11_+_114271367 | 0.28 |
ENST00000544582.1
ENST00000545678.1 |
RBM7
|
RNA binding motif protein 7 |
| chr5_-_75013193 | 0.26 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr17_-_63822563 | 0.25 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr12_-_10826612 | 0.25 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr19_-_58459039 | 0.25 |
ENST00000282308.3
ENST00000598928.1 |
ZNF256
|
zinc finger protein 256 |
| chr19_+_535835 | 0.25 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr22_-_36635598 | 0.24 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr2_+_64068116 | 0.24 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_-_74648702 | 0.23 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr17_+_7344057 | 0.23 |
ENST00000575398.1
ENST00000575082.1 |
FGF11
|
fibroblast growth factor 11 |
| chr9_-_138391692 | 0.23 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr6_+_80816342 | 0.23 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr1_-_24438664 | 0.23 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chr4_-_170924888 | 0.23 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_+_16254488 | 0.23 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr12_+_48592134 | 0.23 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr1_-_1763721 | 0.22 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr4_+_26585538 | 0.22 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr15_+_23810853 | 0.21 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr11_+_45168182 | 0.21 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr6_-_159420780 | 0.21 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr10_-_4720301 | 0.21 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr6_-_90529418 | 0.20 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr1_-_54872059 | 0.20 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr19_-_23578220 | 0.20 |
ENST00000595533.1
ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91
|
zinc finger protein 91 |
| chr4_+_156775910 | 0.19 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_-_4689649 | 0.19 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr7_+_40174565 | 0.19 |
ENST00000309930.5
ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10
|
succinylCoA:glutarate-CoA transferase |
| chr12_-_57030096 | 0.18 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr19_-_30199516 | 0.18 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr19_+_44645700 | 0.18 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr12_+_32655110 | 0.18 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_-_111092873 | 0.18 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr19_+_5914213 | 0.18 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr7_+_70229899 | 0.18 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr5_-_74162153 | 0.18 |
ENST00000514200.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr11_-_60010556 | 0.18 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr14_+_58894404 | 0.17 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr7_+_44040488 | 0.17 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr2_+_102953608 | 0.17 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr16_-_28608364 | 0.17 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr8_-_91657740 | 0.17 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr1_-_110613276 | 0.17 |
ENST00000369792.4
|
ALX3
|
ALX homeobox 3 |
| chr20_-_23807358 | 0.16 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr9_-_26892765 | 0.16 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr19_-_40896081 | 0.16 |
ENST00000291823.2
|
HIPK4
|
homeodomain interacting protein kinase 4 |
| chr12_+_50794947 | 0.16 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr15_+_58724184 | 0.15 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr12_+_57522692 | 0.15 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr11_-_125932685 | 0.15 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr5_-_99870932 | 0.15 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr12_-_75784669 | 0.15 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr7_+_1084206 | 0.15 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr11_-_108338218 | 0.15 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr3_+_197687071 | 0.14 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chrX_-_73512177 | 0.14 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr3_-_141944398 | 0.14 |
ENST00000544571.1
ENST00000392993.2 |
GK5
|
glycerol kinase 5 (putative) |
| chr17_-_4642429 | 0.14 |
ENST00000573123.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr3_-_176914191 | 0.14 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr12_-_91573132 | 0.13 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr12_-_123215306 | 0.13 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr15_-_59225844 | 0.13 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr19_+_5690207 | 0.13 |
ENST00000347512.3
|
RPL36
|
ribosomal protein L36 |
| chr6_-_31088214 | 0.13 |
ENST00000376288.2
|
CDSN
|
corneodesmosin |
| chr2_+_242913327 | 0.13 |
ENST00000426962.1
|
AC093642.3
|
AC093642.3 |
| chrX_-_55187531 | 0.12 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr4_+_160203650 | 0.12 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_153433120 | 0.12 |
ENST00000368723.3
|
S100A7
|
S100 calcium binding protein A7 |
| chr22_+_23029188 | 0.12 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr15_-_100258029 | 0.12 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr8_-_27469383 | 0.12 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr5_-_42811986 | 0.12 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr10_-_4720333 | 0.12 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr8_-_145642203 | 0.12 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr12_+_45609797 | 0.11 |
ENST00000425752.2
|
ANO6
|
anoctamin 6 |
| chr11_+_121447469 | 0.11 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr12_+_133613937 | 0.11 |
ENST00000539354.1
ENST00000542874.1 ENST00000438628.2 |
ZNF84
|
zinc finger protein 84 |
| chr11_-_8795787 | 0.11 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr19_+_5690297 | 0.11 |
ENST00000582463.1
ENST00000579446.1 ENST00000394580.2 |
RPL36
|
ribosomal protein L36 |
| chr20_+_61867235 | 0.11 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr17_+_30771279 | 0.11 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr7_+_151791074 | 0.11 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr17_-_4689727 | 0.11 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chrX_+_129535937 | 0.11 |
ENST00000305536.6
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein, X-linked 2 |
| chr3_+_172034218 | 0.11 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr12_-_57522813 | 0.10 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr21_-_47352477 | 0.10 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr11_-_914774 | 0.10 |
ENST00000528154.1
ENST00000525840.1 |
CHID1
|
chitinase domain containing 1 |
| chr1_-_207095324 | 0.10 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr16_+_22517166 | 0.10 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr7_-_17980091 | 0.10 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr12_+_57522439 | 0.10 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr19_+_44645731 | 0.10 |
ENST00000426739.2
|
ZNF234
|
zinc finger protein 234 |
| chr15_-_59225758 | 0.10 |
ENST00000558486.1
ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM
|
SAFB-like, transcription modulator |
| chr1_+_196788887 | 0.10 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr6_-_109330702 | 0.10 |
ENST00000356644.7
|
SESN1
|
sestrin 1 |
| chr12_-_80328905 | 0.10 |
ENST00000547330.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr5_+_140579162 | 0.10 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr1_-_177939041 | 0.10 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr3_+_99833755 | 0.10 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr3_+_142720366 | 0.10 |
ENST00000493782.1
ENST00000397933.2 ENST00000473835.2 ENST00000493598.2 |
U2SURP
|
U2 snRNP-associated SURP domain containing |
| chr5_-_176057518 | 0.10 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr7_-_22539771 | 0.10 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr12_+_64845864 | 0.10 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr19_-_53606604 | 0.10 |
ENST00000599056.1
ENST00000599247.1 ENST00000355147.5 ENST00000429604.1 ENST00000418871.1 ENST00000599637.1 |
ZNF160
|
zinc finger protein 160 |
| chr16_-_4838255 | 0.09 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr19_-_52531600 | 0.09 |
ENST00000356322.6
ENST00000270649.6 |
ZNF614
|
zinc finger protein 614 |
| chr6_+_31637944 | 0.09 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr17_+_66509019 | 0.09 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr18_-_18691739 | 0.09 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr11_-_85376121 | 0.09 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr11_+_133902547 | 0.09 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr6_-_88299678 | 0.09 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr2_+_95831529 | 0.09 |
ENST00000295210.6
ENST00000453539.2 |
ZNF2
|
zinc finger protein 2 |
| chr12_+_50794891 | 0.09 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr3_-_119379427 | 0.09 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr1_+_17248418 | 0.09 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr13_-_53024725 | 0.09 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr17_-_79304150 | 0.09 |
ENST00000574093.1
|
TMEM105
|
transmembrane protein 105 |
| chr4_-_668108 | 0.09 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr22_+_21133469 | 0.09 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr7_-_30518304 | 0.08 |
ENST00000423334.2
ENST00000411552.1 ENST00000419799.1 |
NOD1
|
nucleotide-binding oligomerization domain containing 1 |
| chr7_+_2695726 | 0.08 |
ENST00000429448.1
|
TTYH3
|
tweety family member 3 |
| chr1_+_81106951 | 0.08 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr4_+_8183793 | 0.08 |
ENST00000509119.1
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr13_-_52733670 | 0.08 |
ENST00000550841.1
|
NEK3
|
NIMA-related kinase 3 |
| chr16_-_3350614 | 0.08 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr10_+_24498060 | 0.08 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr14_+_58894103 | 0.08 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr8_+_19796381 | 0.08 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr11_-_94965667 | 0.08 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr11_-_10879593 | 0.08 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr10_+_38383255 | 0.08 |
ENST00000351773.3
ENST00000361085.5 |
ZNF37A
|
zinc finger protein 37A |
| chr15_+_59499031 | 0.08 |
ENST00000307144.4
|
LDHAL6B
|
lactate dehydrogenase A-like 6B |
| chr2_+_201981527 | 0.08 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_+_32655048 | 0.08 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr4_+_26585686 | 0.08 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr4_-_185395191 | 0.08 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr9_-_125240235 | 0.08 |
ENST00000259357.2
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1 |
| chr5_+_176449684 | 0.08 |
ENST00000506693.1
ENST00000358149.3 ENST00000512315.1 ENST00000503425.1 |
ZNF346
|
zinc finger protein 346 |
| chr10_+_26727125 | 0.08 |
ENST00000376236.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr3_-_119379719 | 0.08 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr5_-_171404730 | 0.08 |
ENST00000518752.1
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr5_-_99870890 | 0.08 |
ENST00000499025.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr18_-_12702711 | 0.08 |
ENST00000423709.2
|
CEP76
|
centrosomal protein 76kDa |
| chr11_+_74811578 | 0.08 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr18_+_72166564 | 0.08 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr1_-_153538292 | 0.08 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr5_-_90610200 | 0.07 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr4_-_175204765 | 0.07 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr8_+_97773202 | 0.07 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr1_-_67600639 | 0.07 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr6_+_88299833 | 0.07 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr1_-_866445 | 0.07 |
ENST00000598827.1
|
AL645608.1
|
Uncharacterized protein |
| chr5_-_137911049 | 0.07 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr2_+_128177253 | 0.07 |
ENST00000427769.1
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr16_+_4838412 | 0.07 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr3_+_121311966 | 0.07 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr11_+_76839283 | 0.07 |
ENST00000409709.3
ENST00000409893.1 ENST00000458637.2 ENST00000409619.2 |
MYO7A
|
myosin VIIA |
| chr7_-_92219698 | 0.07 |
ENST00000438306.1
ENST00000445716.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr21_+_43919710 | 0.07 |
ENST00000398341.3
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr21_+_42733870 | 0.07 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr18_+_61442629 | 0.07 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr16_+_82068585 | 0.07 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr18_+_9913977 | 0.07 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr2_-_167232484 | 0.07 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr8_-_121825088 | 0.07 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr8_-_95220775 | 0.07 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr11_+_72983246 | 0.07 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_-_49198216 | 0.07 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr9_+_103189405 | 0.07 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr12_+_56075330 | 0.07 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr16_+_640201 | 0.07 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr18_-_55253830 | 0.07 |
ENST00000591215.1
|
FECH
|
ferrochelatase |
| chr21_-_30257669 | 0.07 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr6_-_127840336 | 0.07 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr1_+_17516275 | 0.07 |
ENST00000412427.1
|
RP11-380J14.1
|
RP11-380J14.1 |
| chr17_+_20483037 | 0.07 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.4 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.0 | GO:1903524 | positive regulation of blood circulation(GO:1903524) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |