Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
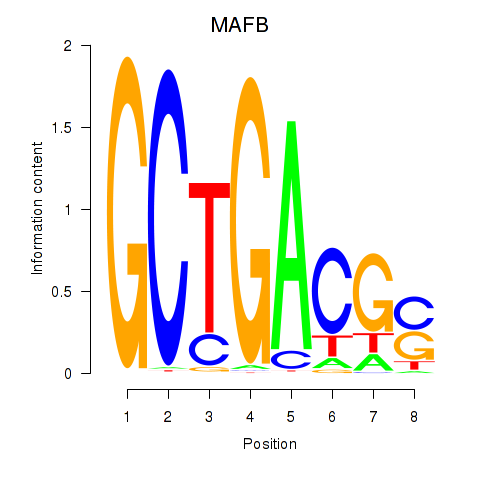
Results for MAFB
Z-value: 1.11
Transcription factors associated with MAFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFB
|
ENSG00000204103.2 | MAF bZIP transcription factor B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFB | hg19_v2_chr20_-_39317868_39317884 | -0.23 | 7.7e-01 | Click! |
Activity profile of MAFB motif
Sorted Z-values of MAFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_57466629 | 0.87 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr7_+_7222233 | 0.62 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr19_+_3224700 | 0.45 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr15_+_40331456 | 0.42 |
ENST00000504245.1
ENST00000560341.1 |
SRP14-AS1
|
SRP14 antisense RNA1 (head to head) |
| chr16_+_226658 | 0.40 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr11_-_46142615 | 0.40 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_30771279 | 0.37 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr14_+_32546485 | 0.37 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr16_+_222846 | 0.36 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr13_-_44735393 | 0.35 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr2_+_191745560 | 0.34 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr21_+_44394742 | 0.34 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr2_-_55277692 | 0.34 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr5_+_61602236 | 0.32 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr3_-_71803474 | 0.31 |
ENST00000448225.1
ENST00000496214.2 |
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr1_+_43148625 | 0.31 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr7_-_27239703 | 0.30 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr2_-_37193606 | 0.30 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr2_-_55277436 | 0.30 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr22_+_31199037 | 0.29 |
ENST00000424224.1
|
OSBP2
|
oxysterol binding protein 2 |
| chr13_+_98086445 | 0.29 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr8_-_92053042 | 0.29 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chr19_+_35773242 | 0.28 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr1_+_78354175 | 0.28 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr14_+_103801140 | 0.28 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr16_+_56965960 | 0.28 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chrY_+_2803322 | 0.28 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr19_-_13068012 | 0.28 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr3_+_37284668 | 0.28 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr19_-_10305302 | 0.28 |
ENST00000592054.1
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr7_+_77167376 | 0.28 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_-_48016023 | 0.27 |
ENST00000598615.1
ENST00000597118.1 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr14_-_50087312 | 0.27 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like |
| chr19_-_39735646 | 0.26 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr17_-_56065484 | 0.26 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr20_-_32700075 | 0.26 |
ENST00000374980.2
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
| chr17_-_4269768 | 0.26 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr1_+_43148059 | 0.26 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr17_-_56065540 | 0.26 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr15_+_27112380 | 0.26 |
ENST00000554596.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr6_+_150070857 | 0.25 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr12_-_76478386 | 0.25 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr14_+_32546274 | 0.25 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr10_+_17686193 | 0.25 |
ENST00000377500.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr2_-_65357225 | 0.25 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr8_+_66556936 | 0.25 |
ENST00000262146.4
|
MTFR1
|
mitochondrial fission regulator 1 |
| chr5_+_61601965 | 0.24 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr8_-_25315905 | 0.24 |
ENST00000221200.4
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr11_+_10772847 | 0.24 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr19_+_39759154 | 0.24 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr12_-_120805872 | 0.24 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr10_+_32735030 | 0.24 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr1_+_161494036 | 0.24 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr5_-_36152031 | 0.24 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr16_+_23847339 | 0.24 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr19_+_55795493 | 0.23 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr20_-_524362 | 0.23 |
ENST00000460062.2
ENST00000608066.1 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr4_+_166128836 | 0.23 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr3_+_31574189 | 0.23 |
ENST00000295770.2
|
STT3B
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr3_-_150481218 | 0.23 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr5_-_118324200 | 0.23 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr6_+_32937083 | 0.23 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr14_+_32546145 | 0.23 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr8_+_57124245 | 0.23 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr9_+_117350009 | 0.23 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr10_+_14880364 | 0.23 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr12_-_93323013 | 0.22 |
ENST00000322349.8
|
EEA1
|
early endosome antigen 1 |
| chr5_+_49962772 | 0.22 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chrX_-_43832711 | 0.22 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr3_-_185542761 | 0.22 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr7_+_6414128 | 0.22 |
ENST00000348035.4
ENST00000356142.4 |
RAC1
|
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| chr15_+_83776324 | 0.22 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr5_-_156390230 | 0.22 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr18_-_77793891 | 0.22 |
ENST00000592957.1
ENST00000585474.1 |
TXNL4A
|
thioredoxin-like 4A |
| chr2_+_191745535 | 0.21 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr3_+_16926441 | 0.21 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr3_+_63897605 | 0.21 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr7_+_107204389 | 0.21 |
ENST00000265720.3
ENST00000402620.1 |
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr12_-_76478417 | 0.21 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr11_+_43702236 | 0.21 |
ENST00000531185.1
ENST00000278353.4 |
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr15_-_40331356 | 0.21 |
ENST00000560773.1
ENST00000267884.6 |
SRP14
|
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr9_+_108210279 | 0.21 |
ENST00000374716.4
ENST00000374710.3 ENST00000481272.1 ENST00000484973.1 ENST00000394926.3 ENST00000539376.1 |
FSD1L
|
fibronectin type III and SPRY domain containing 1-like |
| chr2_+_74741569 | 0.21 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr6_-_170124027 | 0.21 |
ENST00000366780.4
ENST00000339209.4 |
PHF10
|
PHD finger protein 10 |
| chr11_+_10772534 | 0.20 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr7_+_77167343 | 0.20 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chrX_-_83442915 | 0.20 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr7_-_151433342 | 0.20 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr22_-_28315115 | 0.20 |
ENST00000455418.3
ENST00000436663.1 ENST00000320996.10 ENST00000335272.5 |
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr4_-_165305086 | 0.20 |
ENST00000507270.1
ENST00000514618.1 ENST00000503008.1 |
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_-_166136187 | 0.20 |
ENST00000338353.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr11_+_13690249 | 0.20 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr1_+_60280458 | 0.20 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chrX_+_118892545 | 0.19 |
ENST00000343905.3
|
SOWAHD
|
sosondowah ankyrin repeat domain family member D |
| chr1_-_67390474 | 0.19 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr17_-_31620006 | 0.19 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr2_-_157189180 | 0.19 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr9_-_127905736 | 0.19 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr17_-_48943706 | 0.19 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chrX_-_71497148 | 0.19 |
ENST00000316084.6
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr12_+_64798095 | 0.19 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_+_80351977 | 0.19 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr15_-_40331115 | 0.19 |
ENST00000558720.1
|
SRP14
|
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr4_-_89619386 | 0.19 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr12_-_76478446 | 0.19 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr16_-_75529273 | 0.18 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr3_-_113415441 | 0.18 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr9_+_103189458 | 0.18 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr4_-_80994210 | 0.18 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr3_-_167452614 | 0.18 |
ENST00000392750.2
ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10
|
programmed cell death 10 |
| chr4_-_52904425 | 0.18 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr9_+_114393634 | 0.18 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr16_+_28889801 | 0.18 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr4_+_77870960 | 0.18 |
ENST00000505788.1
ENST00000510515.1 ENST00000504637.1 |
SEPT11
|
septin 11 |
| chrX_+_153672468 | 0.18 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chr5_+_178450753 | 0.18 |
ENST00000444149.2
ENST00000519896.1 ENST00000522442.1 |
ZNF879
|
zinc finger protein 879 |
| chr19_-_18314799 | 0.18 |
ENST00000481914.2
|
RAB3A
|
RAB3A, member RAS oncogene family |
| chr16_-_66959429 | 0.18 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr11_-_62572901 | 0.18 |
ENST00000439713.2
ENST00000531131.1 ENST00000530875.1 ENST00000531709.2 ENST00000294172.2 |
NXF1
|
nuclear RNA export factor 1 |
| chr14_-_54955721 | 0.18 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr9_+_128509663 | 0.18 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr11_-_46141338 | 0.18 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr10_-_124768300 | 0.18 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr20_-_524340 | 0.18 |
ENST00000400227.3
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr4_+_190861941 | 0.17 |
ENST00000226798.4
|
FRG1
|
FSHD region gene 1 |
| chr3_-_128712955 | 0.17 |
ENST00000265068.5
|
KIAA1257
|
KIAA1257 |
| chr5_+_61602055 | 0.17 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr2_-_225907150 | 0.17 |
ENST00000258390.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr8_-_80680078 | 0.17 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr11_-_46142948 | 0.17 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr17_+_41150793 | 0.17 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr2_-_174828892 | 0.17 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chr8_-_131028869 | 0.17 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr8_-_102217796 | 0.17 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr10_+_32735177 | 0.17 |
ENST00000545067.1
|
CCDC7
|
coiled-coil domain containing 7 |
| chr3_+_133292851 | 0.17 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr14_+_77843459 | 0.17 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr10_+_91061712 | 0.17 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr7_-_8301768 | 0.17 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr19_-_13617037 | 0.17 |
ENST00000360228.5
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_+_226250379 | 0.17 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr22_+_26565440 | 0.17 |
ENST00000404234.3
ENST00000529632.2 ENST00000360929.3 ENST00000248933.6 ENST00000343706.4 |
SEZ6L
|
seizure related 6 homolog (mouse)-like |
| chr10_+_91087651 | 0.16 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_+_26438289 | 0.16 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr12_-_12509929 | 0.16 |
ENST00000381800.2
|
LOH12CR2
|
loss of heterozygosity, 12, chromosomal region 2 (non-protein coding) |
| chr15_+_83776137 | 0.16 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr7_-_151433393 | 0.16 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr14_-_53258180 | 0.16 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr17_+_66244071 | 0.16 |
ENST00000580548.1
ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr2_-_183903133 | 0.16 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr17_-_72968837 | 0.16 |
ENST00000581676.1
|
HID1
|
HID1 domain containing |
| chr7_+_114562909 | 0.16 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_+_93698974 | 0.16 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr17_-_36413133 | 0.16 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr3_-_71803917 | 0.16 |
ENST00000421769.2
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr11_+_18433840 | 0.16 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr20_+_57466461 | 0.16 |
ENST00000306090.10
|
GNAS
|
GNAS complex locus |
| chr3_+_186288454 | 0.16 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr2_+_71357744 | 0.16 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr3_+_150264458 | 0.16 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr12_+_19593515 | 0.16 |
ENST00000360995.4
|
AEBP2
|
AE binding protein 2 |
| chr2_-_179315490 | 0.16 |
ENST00000487082.1
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr15_+_68346501 | 0.16 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chrX_+_37545012 | 0.16 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr2_+_189156389 | 0.16 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chrX_-_129244336 | 0.16 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chrX_+_135579238 | 0.16 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr5_+_76506706 | 0.16 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr7_+_79764104 | 0.16 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chrX_+_102965835 | 0.16 |
ENST00000319560.6
|
TMEM31
|
transmembrane protein 31 |
| chr17_-_79849438 | 0.16 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr12_-_56709674 | 0.16 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr1_-_40367530 | 0.15 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr8_+_101170257 | 0.15 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr2_-_153573887 | 0.15 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_-_58469687 | 0.15 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr2_+_176957619 | 0.15 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr2_-_56150910 | 0.15 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr7_-_139876734 | 0.15 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr7_-_140178775 | 0.15 |
ENST00000474576.1
ENST00000473444.1 ENST00000471104.1 |
MKRN1
|
makorin ring finger protein 1 |
| chr10_+_17686221 | 0.15 |
ENST00000540523.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr15_+_41624892 | 0.15 |
ENST00000260359.6
ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chr17_-_42276574 | 0.15 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr8_+_87354945 | 0.15 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr8_-_102217515 | 0.15 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr19_+_7587555 | 0.15 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr5_-_95297534 | 0.15 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr1_-_53833812 | 0.15 |
ENST00000449958.1
|
RP11-117D22.2
|
RP11-117D22.2 |
| chr2_-_201828356 | 0.15 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr19_+_1452188 | 0.15 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr3_-_167452703 | 0.15 |
ENST00000497056.2
ENST00000473645.2 |
PDCD10
|
programmed cell death 10 |
| chr7_+_73507409 | 0.15 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr12_+_42624050 | 0.15 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr12_+_50794730 | 0.15 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr12_-_57146095 | 0.15 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr2_-_219925189 | 0.15 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr17_+_8316442 | 0.15 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr10_+_51827648 | 0.15 |
ENST00000351071.6
ENST00000314664.7 ENST00000282633.5 |
FAM21A
|
family with sequence similarity 21, member A |
| chr13_-_52027134 | 0.15 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chrX_+_117629766 | 0.15 |
ENST00000276204.6
ENST00000276202.7 |
DOCK11
|
dedicator of cytokinesis 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.4 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.1 | 1.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.4 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.3 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.3 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 0.5 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.3 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.1 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.2 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.3 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.2 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.2 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.4 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.1 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.4 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:1990927 | vesicle-mediated cholesterol transport(GO:0090119) short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.4 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.3 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.7 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0072038 | mesenchymal stem cell maintenance involved in nephron morphogenesis(GO:0072038) |
| 0.0 | 0.2 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.0 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:0021843 | substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0043103 | hypoxanthine salvage(GO:0043103) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.5 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0042159 | protein depalmitoylation(GO:0002084) lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0070318 | response to anoxia(GO:0034059) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:2000269 | fibroblast apoptotic process(GO:0044346) regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0060018 | positive regulation of chromatin assembly or disassembly(GO:0045799) astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:1902804 | negative regulation of synaptic vesicle transport(GO:1902804) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.2 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.0 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of antigen processing and presentation(GO:0002579) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.0 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.2 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.0 | 0.1 | GO:0050925 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:2000973 | midbrain morphogenesis(GO:1904693) regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0061307 | cardiac neural crest cell differentiation involved in heart development(GO:0061307) cardiac neural crest cell development involved in heart development(GO:0061308) cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.0 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.0 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.0 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.2 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 0.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.0 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.0 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.0 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:1902869 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:1902908 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) regulation of melanosome transport(GO:1902908) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.6 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.0 | GO:0090032 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.4 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0042713 | sperm ejaculation(GO:0042713) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.0 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.0 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.1 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.9 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.2 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0031248 | protein acetyltransferase complex(GO:0031248) acetyltransferase complex(GO:1902493) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.6 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.0 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 1.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0052840 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0050473 | linoleate 13S-lipoxygenase activity(GO:0016165) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) group II metabotropic glutamate receptor activity(GO:0001641) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |