Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for LHX8
Z-value: 0.59
Transcription factors associated with LHX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX8
|
ENSG00000162624.10 | LIM homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX8 | hg19_v2_chr1_+_75594119_75594119 | -0.58 | 4.2e-01 | Click! |
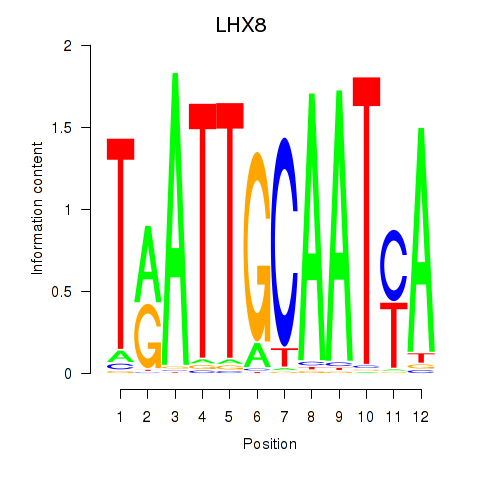
Activity profile of LHX8 motif
Sorted Z-values of LHX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_40074996 | 0.21 |
ENST00000350221.3
|
FSIP1
|
fibrous sheath interacting protein 1 |
| chr4_-_186125077 | 0.20 |
ENST00000458385.2
ENST00000514798.1 ENST00000296775.6 |
KIAA1430
|
KIAA1430 |
| chr16_-_53737722 | 0.20 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chrX_+_108779870 | 0.20 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chrX_+_108779004 | 0.19 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr17_-_57229155 | 0.18 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_-_168464875 | 0.18 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chrX_+_70798261 | 0.18 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr6_+_154360616 | 0.18 |
ENST00000229768.5
ENST00000419506.2 ENST00000524163.1 ENST00000414028.2 ENST00000435918.2 ENST00000337049.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr9_+_26956474 | 0.17 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr2_-_152118276 | 0.17 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr20_+_30458431 | 0.16 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr16_-_15149828 | 0.16 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr17_+_58018269 | 0.16 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr9_+_70856899 | 0.16 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr4_+_76439095 | 0.15 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr14_+_62164340 | 0.15 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr19_-_4535233 | 0.15 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr4_+_174089951 | 0.15 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr2_+_114195268 | 0.15 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr1_+_95616933 | 0.14 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr7_-_120498357 | 0.14 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr3_-_107099454 | 0.14 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chr16_-_53737795 | 0.13 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr2_-_178128250 | 0.13 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr14_-_92247032 | 0.13 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr14_+_57046530 | 0.13 |
ENST00000536419.1
ENST00000538838.1 |
TMEM260
|
transmembrane protein 260 |
| chr6_+_32006042 | 0.13 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr9_-_69262509 | 0.13 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr18_+_71815743 | 0.13 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr19_+_21203426 | 0.13 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr21_-_34863998 | 0.13 |
ENST00000402202.1
ENST00000381947.3 |
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr4_+_57302297 | 0.12 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr11_-_3400442 | 0.12 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr9_-_179018 | 0.12 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr4_+_80413554 | 0.11 |
ENST00000508174.1
|
LINC00989
|
long intergenic non-protein coding RNA 989 |
| chr11_+_101918153 | 0.11 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr13_+_24883703 | 0.11 |
ENST00000332018.4
|
C1QTNF9
|
C1q and tumor necrosis factor related protein 9 |
| chr1_-_236445251 | 0.11 |
ENST00000354619.5
ENST00000327333.8 |
ERO1LB
|
ERO1-like beta (S. cerevisiae) |
| chr1_-_95391315 | 0.11 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr1_+_109419804 | 0.11 |
ENST00000435475.1
|
GPSM2
|
G-protein signaling modulator 2 |
| chr2_-_178128528 | 0.11 |
ENST00000397063.4
ENST00000421929.1 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr16_-_15149917 | 0.11 |
ENST00000287706.3
|
NTAN1
|
N-terminal asparagine amidase |
| chrX_+_84258832 | 0.11 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr1_-_149459549 | 0.11 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr3_+_119187785 | 0.11 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr7_+_151791074 | 0.10 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_-_178128149 | 0.10 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr17_-_76719638 | 0.10 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr12_+_97306295 | 0.10 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr4_-_109684120 | 0.10 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr7_+_7811992 | 0.10 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr1_-_48937838 | 0.10 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr17_+_45000483 | 0.10 |
ENST00000576910.2
ENST00000439730.2 ENST00000393456.2 ENST00000415811.2 ENST00000575949.1 ENST00000225567.4 ENST00000572403.1 ENST00000570879.1 |
GOSR2
|
golgi SNAP receptor complex member 2 |
| chr9_+_26956371 | 0.10 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr2_+_171640291 | 0.10 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr15_-_43785274 | 0.09 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr2_-_136678123 | 0.09 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chrX_+_13707235 | 0.09 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr7_+_30589829 | 0.09 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr10_-_70231639 | 0.09 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr3_+_195447738 | 0.09 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr18_-_74839891 | 0.09 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr3_-_191000172 | 0.09 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr1_+_113009163 | 0.09 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr4_+_174089904 | 0.09 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr16_+_82068585 | 0.09 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr12_-_8088773 | 0.09 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr12_+_48225126 | 0.09 |
ENST00000550909.1
ENST00000550720.1 ENST00000548564.1 |
RP5-1057I20.2
|
RP5-1057I20.2 |
| chr4_+_155484103 | 0.08 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr9_+_70856397 | 0.08 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr10_+_94352956 | 0.08 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr7_-_14026063 | 0.08 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr5_-_75008244 | 0.08 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr7_-_28220354 | 0.08 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr1_+_247670360 | 0.08 |
ENST00000527084.1
ENST00000527541.1 ENST00000366490.3 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr22_+_29876197 | 0.08 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr9_-_20622478 | 0.08 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_+_109237717 | 0.08 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_+_78832747 | 0.08 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr9_-_70490107 | 0.08 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr3_-_112693759 | 0.08 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr5_+_41904431 | 0.07 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr15_+_33022885 | 0.07 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr21_-_16254231 | 0.07 |
ENST00000412426.1
ENST00000418954.1 |
AF127936.7
|
AF127936.7 |
| chr21_-_33984456 | 0.07 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr13_-_24476794 | 0.07 |
ENST00000382140.2
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr1_+_47603109 | 0.07 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr6_+_76599809 | 0.07 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr3_-_48956818 | 0.07 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr7_-_103086598 | 0.07 |
ENST00000339444.6
ENST00000356767.4 ENST00000393735.2 ENST00000306312.3 ENST00000432958.2 |
SLC26A5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr6_-_75828774 | 0.07 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr5_+_135170331 | 0.07 |
ENST00000425402.1
ENST00000274513.5 ENST00000420621.1 ENST00000433282.2 ENST00000412661.2 |
SLC25A48
|
solute carrier family 25, member 48 |
| chr13_+_38923959 | 0.07 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr2_+_128377550 | 0.07 |
ENST00000437387.1
ENST00000409090.1 |
MYO7B
|
myosin VIIB |
| chr12_+_48866448 | 0.06 |
ENST00000266594.1
|
ANP32D
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member D |
| chr1_-_212004090 | 0.06 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr17_-_38821373 | 0.06 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr4_-_83812402 | 0.06 |
ENST00000395310.2
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr5_-_147162263 | 0.06 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr1_+_92632542 | 0.06 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr8_-_62602327 | 0.06 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr14_+_101295638 | 0.06 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr8_+_75262629 | 0.06 |
ENST00000434412.2
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr4_-_83812248 | 0.06 |
ENST00000514326.1
ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr19_-_695427 | 0.06 |
ENST00000329267.7
|
PRSS57
|
protease, serine, 57 |
| chr8_+_17354617 | 0.06 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr1_+_145883868 | 0.06 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr7_+_151791037 | 0.06 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr3_-_112693865 | 0.06 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr10_+_69869237 | 0.05 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr4_+_155484155 | 0.05 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr15_-_35047166 | 0.05 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr13_-_37633567 | 0.05 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr12_+_54410664 | 0.05 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr1_+_66796401 | 0.05 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr7_+_5938386 | 0.05 |
ENST00000537980.1
|
CCZ1
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) |
| chr11_+_59480899 | 0.05 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr8_-_141810634 | 0.05 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_212208919 | 0.05 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr14_+_57046500 | 0.05 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr9_+_96846740 | 0.05 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr3_-_119182523 | 0.05 |
ENST00000319172.5
|
TMEM39A
|
transmembrane protein 39A |
| chr15_-_43785303 | 0.05 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_+_70876926 | 0.05 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr7_-_6865826 | 0.05 |
ENST00000538180.1
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr10_+_90672113 | 0.05 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr1_-_160492994 | 0.05 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr7_+_12727250 | 0.05 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr4_+_26344754 | 0.05 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_86947296 | 0.05 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chrX_+_73164149 | 0.05 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr4_-_69111401 | 0.05 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr8_+_75262612 | 0.05 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr10_-_29923893 | 0.05 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_+_119957554 | 0.04 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr18_-_14132422 | 0.04 |
ENST00000589498.1
ENST00000590202.1 |
ZNF519
|
zinc finger protein 519 |
| chr11_+_47293795 | 0.04 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr11_-_117800080 | 0.04 |
ENST00000524993.1
ENST00000528626.1 ENST00000445164.2 ENST00000430170.2 ENST00000526090.1 |
TMPRSS13
|
transmembrane protease, serine 13 |
| chr4_+_71248795 | 0.04 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr2_-_145275228 | 0.04 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_106631966 | 0.04 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr1_+_224301787 | 0.04 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr6_+_116691001 | 0.04 |
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase |
| chr12_+_122326630 | 0.04 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr12_-_71551652 | 0.04 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_-_149470229 | 0.04 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr4_-_48116540 | 0.04 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr4_-_36245561 | 0.04 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr1_+_165864821 | 0.04 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr2_-_136288113 | 0.04 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_+_87144738 | 0.04 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr1_-_33502528 | 0.04 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chrX_-_23926004 | 0.04 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr11_+_125757556 | 0.04 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr12_+_45686457 | 0.04 |
ENST00000441606.2
|
ANO6
|
anoctamin 6 |
| chr1_+_45241109 | 0.04 |
ENST00000396651.3
|
RPS8
|
ribosomal protein S8 |
| chr6_+_43543942 | 0.04 |
ENST00000372226.1
ENST00000443535.1 |
POLH
|
polymerase (DNA directed), eta |
| chr19_-_58204128 | 0.04 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr2_+_216946589 | 0.04 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chr2_-_178128199 | 0.04 |
ENST00000449627.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr3_-_52804872 | 0.04 |
ENST00000535191.1
ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4
|
NIMA-related kinase 4 |
| chr2_-_145275109 | 0.03 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_-_140482926 | 0.03 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr11_-_72504681 | 0.03 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chrX_-_19689106 | 0.03 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_-_146040968 | 0.03 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr8_+_99076509 | 0.03 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr5_-_39270725 | 0.03 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr4_-_163085107 | 0.03 |
ENST00000379164.4
|
FSTL5
|
follistatin-like 5 |
| chr12_-_8088871 | 0.03 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr13_+_96085847 | 0.03 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chrX_+_73164167 | 0.03 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr19_+_42037433 | 0.03 |
ENST00000599316.1
ENST00000599770.1 |
AC006129.1
|
AC006129.1 |
| chr8_+_17354587 | 0.03 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr11_-_66964638 | 0.03 |
ENST00000444002.2
|
AP001885.1
|
AP001885.1 |
| chr19_+_44716678 | 0.03 |
ENST00000586228.1
ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227
|
zinc finger protein 227 |
| chr19_+_12175504 | 0.03 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr1_-_48937821 | 0.03 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr12_+_26205496 | 0.03 |
ENST00000537946.1
ENST00000541218.1 ENST00000282884.9 ENST00000545413.1 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr12_+_15475331 | 0.03 |
ENST00000281171.4
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr7_+_115862858 | 0.03 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr15_-_44116873 | 0.02 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr2_-_152830479 | 0.02 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr14_-_101295407 | 0.02 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr2_+_234621551 | 0.02 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr6_+_160221293 | 0.02 |
ENST00000610273.1
ENST00000392167.3 |
PNLDC1
|
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
| chr2_+_201994042 | 0.02 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_+_113354341 | 0.02 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chrX_-_20236970 | 0.02 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr2_-_74618964 | 0.02 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr10_-_101825151 | 0.02 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr14_-_36990354 | 0.02 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr4_-_186733363 | 0.02 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_13165457 | 0.02 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr5_-_150473127 | 0.02 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_-_153332886 | 0.02 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr17_-_26220366 | 0.02 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr2_-_50201327 | 0.02 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr15_-_75918787 | 0.02 |
ENST00000564086.1
|
SNUPN
|
snurportin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.2 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0034226 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.1 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:1902990 | DNA replication, removal of RNA primer(GO:0043137) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.3 | GO:0043584 | lateral ventricle development(GO:0021670) nose development(GO:0043584) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.3 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |