Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
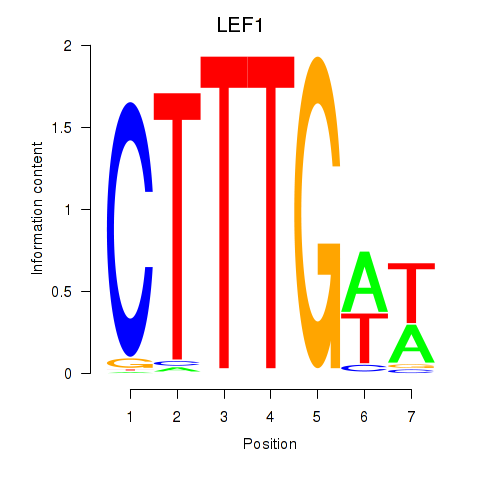
Results for LEF1
Z-value: 0.82
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | lymphoid enhancer binding factor 1 |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_23791484 | 1.03 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr4_+_76649753 | 0.74 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr16_+_53242350 | 0.68 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_110001723 | 0.49 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr17_+_70026795 | 0.43 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr7_-_27219849 | 0.35 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr17_+_76037081 | 0.35 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr5_+_149865377 | 0.35 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_+_12008986 | 0.30 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr22_+_37959647 | 0.29 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr5_+_175288631 | 0.27 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr2_-_97534312 | 0.25 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr12_+_95611536 | 0.25 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr4_+_95972822 | 0.24 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr3_+_182983126 | 0.24 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr10_-_18948208 | 0.24 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr4_+_96012585 | 0.23 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr14_+_64565442 | 0.22 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr9_-_97356075 | 0.20 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr2_-_178129551 | 0.20 |
ENST00000430047.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr4_-_157892055 | 0.20 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr12_+_54378849 | 0.20 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr11_-_94965667 | 0.19 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr17_+_26369865 | 0.19 |
ENST00000582037.1
|
NLK
|
nemo-like kinase |
| chr6_-_127840021 | 0.19 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr1_+_53793885 | 0.19 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr3_+_107244229 | 0.19 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr4_-_140477910 | 0.18 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr3_-_141747950 | 0.18 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_-_139756791 | 0.18 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr17_+_29815013 | 0.18 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr3_-_141747439 | 0.18 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr21_+_17791648 | 0.17 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_+_28724129 | 0.17 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr1_+_244998918 | 0.17 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr8_-_124553437 | 0.17 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr7_+_69064566 | 0.17 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr11_-_85430356 | 0.17 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr13_-_74708372 | 0.16 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr12_+_95612006 | 0.16 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_+_213123862 | 0.16 |
ENST00000366966.2
ENST00000366964.3 |
VASH2
|
vasohibin 2 |
| chr11_-_46141338 | 0.16 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr4_+_24661479 | 0.16 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr2_+_11679963 | 0.16 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chrX_+_78003204 | 0.16 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chrX_+_38420783 | 0.16 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr1_+_93645314 | 0.16 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr6_-_127840336 | 0.16 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr4_-_76649546 | 0.16 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_-_118943930 | 0.15 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr10_+_114710516 | 0.15 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr17_-_77924627 | 0.15 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr15_+_63335899 | 0.15 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr10_+_114710425 | 0.15 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr18_-_74207146 | 0.15 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr6_-_166401442 | 0.15 |
ENST00000584179.1
|
LINC00473
|
long intergenic non-protein coding RNA 473 |
| chr18_-_45456693 | 0.15 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr10_+_114710211 | 0.15 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_196743943 | 0.15 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr6_-_11807277 | 0.15 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_+_174844645 | 0.15 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_-_140178726 | 0.14 |
ENST00000480552.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr9_+_125795788 | 0.14 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr3_+_186648507 | 0.14 |
ENST00000458216.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr12_-_108154705 | 0.14 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chrX_+_100645812 | 0.14 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr7_+_6071007 | 0.14 |
ENST00000409061.1
|
ANKRD61
|
ankyrin repeat domain 61 |
| chr7_+_142982023 | 0.14 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr5_-_16936340 | 0.14 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr1_-_78444776 | 0.14 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr3_-_79068138 | 0.14 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr3_-_52719912 | 0.13 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr1_+_6845384 | 0.13 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr18_+_3450161 | 0.13 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_80548493 | 0.13 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr7_+_20370300 | 0.13 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr7_+_151791037 | 0.13 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr7_+_151791074 | 0.13 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr4_-_140477353 | 0.13 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_+_116451100 | 0.13 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr14_-_51027838 | 0.13 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr4_+_170581213 | 0.13 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr1_-_78444738 | 0.12 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr10_-_21806759 | 0.12 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr15_+_52043813 | 0.12 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr2_-_161056762 | 0.12 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr1_+_93913713 | 0.12 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr12_-_109797249 | 0.12 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr1_+_47799542 | 0.12 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr2_+_162087577 | 0.12 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr8_+_29953163 | 0.12 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chrX_+_86772787 | 0.12 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr1_+_27668505 | 0.12 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr11_-_87908600 | 0.12 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr21_+_18811205 | 0.12 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chr5_-_137514333 | 0.12 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr8_-_42358742 | 0.12 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr12_-_49504449 | 0.12 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr9_-_117150303 | 0.12 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr9_-_117150243 | 0.12 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr8_+_61592073 | 0.12 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr5_-_58571935 | 0.12 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_+_42873548 | 0.12 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr2_+_233734994 | 0.12 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr16_+_53241854 | 0.12 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr2_+_149402989 | 0.11 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr5_+_112074029 | 0.11 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr1_+_197886461 | 0.11 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr6_-_127840048 | 0.11 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr2_+_58655461 | 0.11 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr17_-_47308128 | 0.11 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr4_+_26165074 | 0.11 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_47521309 | 0.11 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr16_+_67063036 | 0.11 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr8_-_22089533 | 0.11 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr17_+_19030782 | 0.11 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr15_+_101402041 | 0.11 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr4_+_86699834 | 0.11 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_-_22089845 | 0.11 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr1_+_78383813 | 0.11 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr11_-_8892464 | 0.10 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr6_+_27925019 | 0.10 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr11_-_133826852 | 0.10 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chrX_-_128788914 | 0.10 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr7_-_27213893 | 0.10 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chrX_+_86772707 | 0.10 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr4_+_96012614 | 0.10 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr11_-_46848393 | 0.10 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr13_-_67802549 | 0.10 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr13_+_39612442 | 0.10 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr2_-_160472952 | 0.10 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_-_112360116 | 0.10 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr10_+_115312766 | 0.10 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr3_-_71179988 | 0.10 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr18_+_55712915 | 0.10 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_-_65018834 | 0.10 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr7_-_27169801 | 0.10 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr13_+_76210448 | 0.10 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr4_-_157892167 | 0.10 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr8_+_21777243 | 0.10 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr3_+_20081515 | 0.10 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr5_-_137514617 | 0.10 |
ENST00000254900.5
|
BRD8
|
bromodomain containing 8 |
| chr2_-_107503558 | 0.10 |
ENST00000361686.4
ENST00000409087.3 |
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr11_-_62432641 | 0.10 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr10_-_14050522 | 0.10 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr4_-_83351294 | 0.10 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr2_+_152214098 | 0.10 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr3_-_71179699 | 0.09 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr1_+_84629976 | 0.09 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr11_-_111649015 | 0.09 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_+_140248518 | 0.09 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr14_-_71107921 | 0.09 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_+_144339738 | 0.09 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr3_+_183353356 | 0.09 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr1_-_23670752 | 0.09 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chrX_-_153095813 | 0.09 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr17_-_44270133 | 0.09 |
ENST00000262419.6
ENST00000393476.3 |
KANSL1
|
KAT8 regulatory NSL complex subunit 1 |
| chr11_-_111649074 | 0.09 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_+_150076406 | 0.09 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr16_+_67062996 | 0.09 |
ENST00000561924.2
|
CBFB
|
core-binding factor, beta subunit |
| chr5_-_137514288 | 0.09 |
ENST00000454473.1
ENST00000418329.1 ENST00000455658.2 ENST00000230901.5 ENST00000402931.1 |
BRD8
|
bromodomain containing 8 |
| chr9_+_36036430 | 0.09 |
ENST00000377966.3
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr15_+_40731920 | 0.09 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr11_-_64013663 | 0.09 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr13_+_76334795 | 0.09 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr2_-_48132814 | 0.08 |
ENST00000316377.4
ENST00000378314.3 |
FBXO11
|
F-box protein 11 |
| chr7_-_84121858 | 0.08 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_7533439 | 0.08 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr11_-_47207390 | 0.08 |
ENST00000539589.1
ENST00000528462.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr6_-_166401527 | 0.08 |
ENST00000455853.1
ENST00000584911.1 |
LINC00473
|
long intergenic non-protein coding RNA 473 |
| chr17_+_75447326 | 0.08 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr5_+_145317356 | 0.08 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr9_+_504674 | 0.08 |
ENST00000382297.2
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr1_+_196621156 | 0.08 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr1_+_12538594 | 0.08 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_-_219615984 | 0.08 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr8_-_95220775 | 0.08 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr17_-_46682321 | 0.08 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr3_+_57875711 | 0.08 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr6_+_108881012 | 0.08 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr6_+_35704855 | 0.08 |
ENST00000288065.2
ENST00000373866.3 |
ARMC12
|
armadillo repeat containing 12 |
| chr22_-_39150947 | 0.08 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr4_+_160203650 | 0.08 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_24645956 | 0.08 |
ENST00000543707.1
|
KIAA0319
|
KIAA0319 |
| chr6_+_36839616 | 0.08 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr10_+_114709999 | 0.08 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_-_74088800 | 0.08 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr12_-_70093235 | 0.08 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr11_-_130184470 | 0.08 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr1_-_202113805 | 0.08 |
ENST00000272217.2
|
ARL8A
|
ADP-ribosylation factor-like 8A |
| chr3_-_33686925 | 0.08 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_139195350 | 0.08 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr6_+_135502501 | 0.08 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr2_-_74780176 | 0.08 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr1_+_84630367 | 0.08 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_27113963 | 0.08 |
ENST00000430292.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr9_+_97766409 | 0.08 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr2_-_86564696 | 0.07 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr1_+_180199393 | 0.07 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr9_+_128510454 | 0.07 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.5 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.2 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.3 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.0 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.4 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.0 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.0 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |