Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
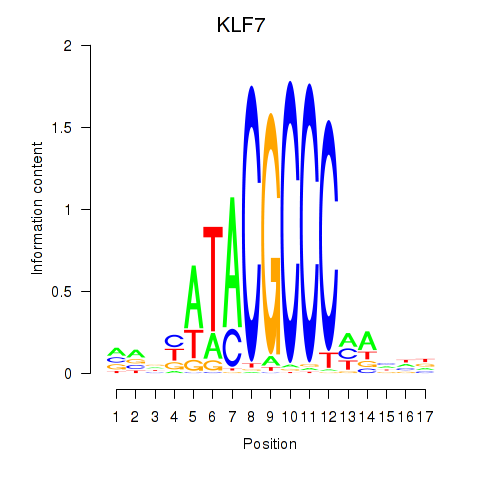
Results for KLF7
Z-value: 0.35
Transcription factors associated with KLF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF7
|
ENSG00000118263.10 | Kruppel like factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF7 | hg19_v2_chr2_-_208030295_208030332 | -0.74 | 2.6e-01 | Click! |
Activity profile of KLF7 motif
Sorted Z-values of KLF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_19281787 | 0.13 |
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr12_-_49581152 | 0.12 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_157108266 | 0.12 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr12_-_49582593 | 0.12 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr17_+_38375528 | 0.11 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr19_-_39390440 | 0.10 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr15_+_96873921 | 0.09 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_30585486 | 0.09 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr19_-_39390350 | 0.08 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr1_-_19229218 | 0.08 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr19_-_7694417 | 0.08 |
ENST00000358368.4
ENST00000534844.1 |
XAB2
|
XPA binding protein 2 |
| chr20_-_2821271 | 0.08 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr12_-_49582837 | 0.08 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr17_-_18218237 | 0.08 |
ENST00000542570.1
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr20_-_2821756 | 0.07 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr16_+_81772633 | 0.07 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr6_+_43603552 | 0.06 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr17_+_41476327 | 0.06 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr17_-_18218270 | 0.06 |
ENST00000321105.5
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr1_-_19229014 | 0.05 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr19_-_40931891 | 0.05 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr19_+_39390320 | 0.05 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_-_74710078 | 0.05 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr9_+_130565147 | 0.05 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr1_-_19229248 | 0.05 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr8_+_72587535 | 0.05 |
ENST00000519840.1
ENST00000521131.1 |
RP11-1144P22.1
|
RP11-1144P22.1 |
| chr22_-_30987849 | 0.04 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr22_-_30987837 | 0.04 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr2_+_113479063 | 0.04 |
ENST00000327581.4
|
NT5DC4
|
5'-nucleotidase domain containing 4 |
| chr1_-_6453399 | 0.04 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr19_+_39390587 | 0.04 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr8_+_17104401 | 0.04 |
ENST00000324815.3
ENST00000518038.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr1_-_21948906 | 0.03 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr5_+_141348598 | 0.03 |
ENST00000394520.2
ENST00000347642.3 |
RNF14
|
ring finger protein 14 |
| chr19_-_14629224 | 0.03 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr5_+_141348721 | 0.03 |
ENST00000507163.1
ENST00000394519.1 |
RNF14
|
ring finger protein 14 |
| chr9_+_130565487 | 0.02 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr11_+_18417948 | 0.02 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr17_-_48277552 | 0.02 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr1_-_6453426 | 0.02 |
ENST00000545482.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr12_+_1099675 | 0.02 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr17_+_38375574 | 0.01 |
ENST00000323571.4
ENST00000585043.1 ENST00000394103.3 ENST00000536600.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr3_-_33138624 | 0.01 |
ENST00000445488.2
ENST00000307377.8 ENST00000440656.1 ENST00000436768.1 ENST00000307363.5 |
GLB1
|
galactosidase, beta 1 |
| chr16_+_20499024 | 0.01 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr6_-_33239612 | 0.01 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr6_+_33239787 | 0.01 |
ENST00000439602.2
ENST00000474973.1 |
RPS18
|
ribosomal protein S18 |
| chr6_-_30585009 | 0.01 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
| chr2_+_74710194 | 0.01 |
ENST00000410003.1
ENST00000442235.2 ENST00000233623.5 |
TTC31
|
tetratricopeptide repeat domain 31 |
| chr20_+_2821366 | 0.01 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr15_+_92937058 | 0.01 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr1_-_161102421 | 0.00 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chrX_-_92928557 | 0.00 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr14_+_29234870 | 0.00 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr9_+_130565568 | 0.00 |
ENST00000423577.1
|
FPGS
|
folylpolyglutamate synthase |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.2 | GO:0019470 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.1 | 0.2 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |