Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for KLF6
Z-value: 0.21
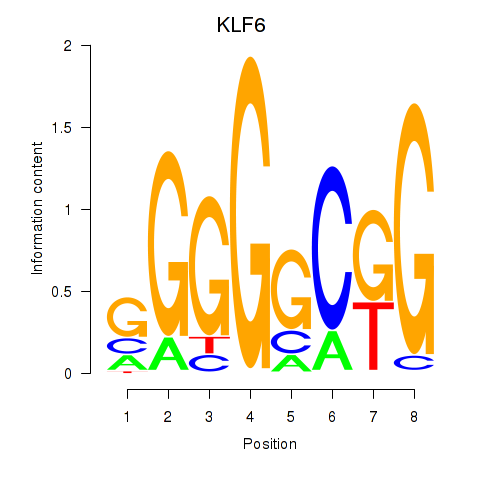
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.10 | Kruppel like factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg19_v2_chr10_-_3827371_3827386 | 0.70 | 3.0e-01 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_110500826 | 0.11 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr3_-_113415441 | 0.11 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr12_-_80328700 | 0.08 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr17_-_27277615 | 0.07 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr8_-_101322132 | 0.07 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr11_+_64009072 | 0.07 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr10_+_76586348 | 0.06 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr4_+_183370146 | 0.06 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr18_-_51750948 | 0.06 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr8_-_125486755 | 0.06 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr10_+_70587279 | 0.06 |
ENST00000298596.6
ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1
|
storkhead box 1 |
| chr7_-_72936608 | 0.06 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr11_-_65640071 | 0.06 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr11_+_64008525 | 0.06 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr11_-_65429891 | 0.06 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr1_-_114696472 | 0.06 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr12_+_69004805 | 0.06 |
ENST00000541216.1
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr11_-_61196858 | 0.06 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr17_+_42081914 | 0.05 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr22_+_35653445 | 0.05 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr6_+_12012170 | 0.05 |
ENST00000487103.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr3_+_133292759 | 0.05 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chrX_+_69674943 | 0.05 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr5_-_81046904 | 0.05 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr11_+_65686728 | 0.05 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr7_-_72936531 | 0.05 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr7_-_130353553 | 0.05 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr20_+_31350184 | 0.05 |
ENST00000328111.2
ENST00000353855.2 ENST00000348286.2 |
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta |
| chr4_+_89444961 | 0.05 |
ENST00000513325.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr3_-_171178157 | 0.05 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr1_-_23670752 | 0.05 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr15_+_44084040 | 0.05 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr17_-_58469329 | 0.05 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr10_-_112678904 | 0.05 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr3_+_97540876 | 0.05 |
ENST00000419587.2
|
CRYBG3
|
Beta/gamma crystallin domain-containing protein 3; cDNA FLJ60082, weakly similar to Uro-adherence factor A |
| chr5_-_168006324 | 0.05 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr13_+_103249322 | 0.05 |
ENST00000376065.4
ENST00000376052.3 |
TPP2
|
tripeptidyl peptidase II |
| chr12_-_57824561 | 0.05 |
ENST00000448732.1
|
R3HDM2
|
R3H domain containing 2 |
| chr1_-_23670813 | 0.05 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr9_+_6645887 | 0.04 |
ENST00000413145.1
|
RP11-390F4.6
|
RP11-390F4.6 |
| chr1_+_43148625 | 0.04 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr9_-_117267717 | 0.04 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr2_-_39347524 | 0.04 |
ENST00000395038.2
ENST00000402219.2 |
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr20_+_57466629 | 0.04 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr6_-_110501126 | 0.04 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr13_-_52027134 | 0.04 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr17_+_30771279 | 0.04 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr16_-_49315731 | 0.04 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr5_+_40679584 | 0.04 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr3_-_112280709 | 0.04 |
ENST00000402314.2
ENST00000283290.5 ENST00000492886.1 |
ATG3
|
autophagy related 3 |
| chr4_+_103790462 | 0.04 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr3_-_167813132 | 0.04 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr15_-_72410455 | 0.04 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr6_-_150039170 | 0.04 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr11_-_94706705 | 0.04 |
ENST00000279839.6
|
CWC15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr19_+_10400615 | 0.04 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr7_-_27239703 | 0.04 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr7_+_86781778 | 0.04 |
ENST00000432937.2
|
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr9_-_115249484 | 0.04 |
ENST00000457681.1
|
C9orf147
|
chromosome 9 open reading frame 147 |
| chr11_+_129939779 | 0.04 |
ENST00000533195.1
ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr1_+_63788730 | 0.04 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr5_+_67511524 | 0.04 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_+_65686952 | 0.04 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr4_-_78740769 | 0.04 |
ENST00000512485.1
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr1_-_23670817 | 0.04 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr13_-_27745936 | 0.04 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr15_-_75743915 | 0.04 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr17_-_36413133 | 0.04 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr14_-_73925225 | 0.04 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr8_-_101963482 | 0.04 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_-_71842706 | 0.04 |
ENST00000563104.1
ENST00000569975.1 ENST00000565412.1 ENST00000567583.1 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr14_+_23790690 | 0.04 |
ENST00000556821.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr10_-_75634219 | 0.04 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr10_+_76871229 | 0.04 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr10_-_75634326 | 0.04 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr11_+_65687158 | 0.04 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr11_+_57365150 | 0.04 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr2_-_61245363 | 0.03 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr1_+_112939121 | 0.03 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr6_-_94129244 | 0.03 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr7_+_70229899 | 0.03 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr19_+_10196981 | 0.03 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr17_-_79008373 | 0.03 |
ENST00000577066.1
ENST00000573167.1 |
BAIAP2-AS1
|
BAIAP2 antisense RNA 1 (head to head) |
| chr10_-_32217717 | 0.03 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chrX_-_118986911 | 0.03 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr3_-_51533966 | 0.03 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr22_+_38004942 | 0.03 |
ENST00000439161.1
ENST00000449944.1 ENST00000411501.1 ENST00000453208.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr8_+_67976593 | 0.03 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr4_-_103789996 | 0.03 |
ENST00000508238.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_111746966 | 0.03 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr19_+_38880695 | 0.03 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_-_51611477 | 0.03 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr4_-_103790026 | 0.03 |
ENST00000338145.3
ENST00000357194.6 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_+_65686802 | 0.03 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr2_+_203499901 | 0.03 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr4_-_76598326 | 0.03 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr8_-_38034234 | 0.03 |
ENST00000311351.4
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr20_-_524340 | 0.03 |
ENST00000400227.3
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr14_-_73924954 | 0.03 |
ENST00000555307.1
ENST00000554818.1 |
NUMB
|
numb homolog (Drosophila) |
| chr14_-_92572894 | 0.03 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr18_-_23670546 | 0.03 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr9_+_130478345 | 0.03 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr14_+_58765305 | 0.03 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr12_+_79258444 | 0.03 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr5_+_60628074 | 0.03 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr2_-_160472052 | 0.03 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr20_-_524362 | 0.03 |
ENST00000460062.2
ENST00000608066.1 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr17_-_58469687 | 0.03 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr16_+_53164833 | 0.03 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr18_-_51751132 | 0.03 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr7_-_143105941 | 0.03 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr9_+_129987488 | 0.03 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr20_-_524415 | 0.03 |
ENST00000400217.2
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr14_-_99947121 | 0.03 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chrX_+_21959108 | 0.03 |
ENST00000457085.1
|
SMS
|
spermine synthase |
| chr5_+_109025067 | 0.03 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr14_+_23791159 | 0.03 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr15_+_64388166 | 0.03 |
ENST00000353874.4
ENST00000261889.5 ENST00000559844.1 ENST00000561026.1 ENST00000558040.1 |
SNX1
|
sorting nexin 1 |
| chr17_+_7341586 | 0.03 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr10_-_75255724 | 0.03 |
ENST00000342558.3
ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr4_-_153456153 | 0.03 |
ENST00000603548.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr14_+_89029336 | 0.03 |
ENST00000556945.1
ENST00000556158.1 ENST00000557607.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr1_-_236445251 | 0.03 |
ENST00000354619.5
ENST00000327333.8 |
ERO1LB
|
ERO1-like beta (S. cerevisiae) |
| chr10_+_112631699 | 0.03 |
ENST00000444997.1
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr17_+_34136459 | 0.03 |
ENST00000588240.1
ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr6_-_30709980 | 0.03 |
ENST00000416018.1
ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1
|
flotillin 1 |
| chr14_-_82000140 | 0.03 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr10_+_114709999 | 0.03 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr17_+_41177220 | 0.03 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr11_+_71259466 | 0.03 |
ENST00000528743.2
|
KRTAP5-9
|
keratin associated protein 5-9 |
| chr16_+_69599899 | 0.03 |
ENST00000567239.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr10_-_112678976 | 0.03 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr6_-_35656685 | 0.03 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr12_+_14518598 | 0.03 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_220306745 | 0.03 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr6_-_131384373 | 0.03 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr12_+_79258547 | 0.03 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr8_-_57123815 | 0.03 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr12_+_56473628 | 0.03 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_-_182360498 | 0.03 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr2_+_219081817 | 0.03 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr17_-_42994283 | 0.03 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr13_+_28712614 | 0.03 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_-_47205457 | 0.03 |
ENST00000409792.3
|
SETD2
|
SET domain containing 2 |
| chr1_+_40506255 | 0.03 |
ENST00000421589.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr2_+_170590321 | 0.03 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr1_+_43148059 | 0.03 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr16_-_14724131 | 0.03 |
ENST00000341484.7
|
PARN
|
poly(A)-specific ribonuclease |
| chr5_-_81046841 | 0.03 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr3_-_88108212 | 0.03 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr15_-_43802769 | 0.03 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr15_+_66161802 | 0.03 |
ENST00000566233.1
ENST00000565075.1 ENST00000435304.2 |
RAB11A
|
RAB11A, member RAS oncogene family |
| chr20_-_40247133 | 0.03 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr17_-_44270133 | 0.03 |
ENST00000262419.6
ENST00000393476.3 |
KANSL1
|
KAT8 regulatory NSL complex subunit 1 |
| chr1_+_207226574 | 0.03 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr20_-_35724388 | 0.03 |
ENST00000344359.3
ENST00000373664.3 |
RBL1
|
retinoblastoma-like 1 (p107) |
| chr2_-_219433014 | 0.02 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr2_+_111878483 | 0.02 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr14_+_89029253 | 0.02 |
ENST00000251038.5
ENST00000359301.3 ENST00000302216.8 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr17_-_53499218 | 0.02 |
ENST00000571578.1
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr3_+_52280220 | 0.02 |
ENST00000409502.3
ENST00000323588.4 |
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr17_+_30264014 | 0.02 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr9_-_120177342 | 0.02 |
ENST00000361209.2
|
ASTN2
|
astrotactin 2 |
| chr2_+_42795745 | 0.02 |
ENST00000406911.1
|
MTA3
|
metastasis associated 1 family, member 3 |
| chr7_-_99774945 | 0.02 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr2_-_159313214 | 0.02 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr4_+_103790120 | 0.02 |
ENST00000273986.4
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr7_+_114562909 | 0.02 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr5_-_159546396 | 0.02 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr19_+_55795493 | 0.02 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr9_+_6413317 | 0.02 |
ENST00000276893.5
ENST00000381373.3 |
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr18_+_9913977 | 0.02 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr3_-_24536453 | 0.02 |
ENST00000453729.2
ENST00000413780.1 |
THRB
|
thyroid hormone receptor, beta |
| chr6_+_110501621 | 0.02 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr10_-_104474128 | 0.02 |
ENST00000260746.5
|
ARL3
|
ADP-ribosylation factor-like 3 |
| chr10_-_124768300 | 0.02 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr9_+_100745615 | 0.02 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr2_-_27341765 | 0.02 |
ENST00000405600.1
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr3_-_167813672 | 0.02 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr2_-_39348137 | 0.02 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr2_+_159313452 | 0.02 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr11_-_94964210 | 0.02 |
ENST00000416495.2
ENST00000393234.1 |
SESN3
|
sestrin 3 |
| chr6_-_35656712 | 0.02 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr9_-_132597529 | 0.02 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr1_-_155243235 | 0.02 |
ENST00000355560.4
ENST00000368361.4 |
CLK2
|
CDC-like kinase 2 |
| chr15_+_41952591 | 0.02 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr15_+_52311398 | 0.02 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr14_-_39572345 | 0.02 |
ENST00000548032.2
ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A
|
Sec23 homolog A (S. cerevisiae) |
| chr7_+_86781847 | 0.02 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr14_-_62162541 | 0.02 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr6_+_110501344 | 0.02 |
ENST00000368932.1
|
CDC40
|
cell division cycle 40 |
| chr11_+_75526212 | 0.02 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr18_-_12884150 | 0.02 |
ENST00000591115.1
ENST00000309660.5 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr4_-_78740511 | 0.02 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr1_-_182360918 | 0.02 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr6_-_170124027 | 0.02 |
ENST00000366780.4
ENST00000339209.4 |
PHF10
|
PHD finger protein 10 |
| chr2_-_161349909 | 0.02 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr12_+_118814185 | 0.02 |
ENST00000543473.1
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr11_-_62521614 | 0.02 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr7_+_86781677 | 0.02 |
ENST00000331242.7
ENST00000394702.3 ENST00000413276.2 ENST00000446796.2 ENST00000411766.2 ENST00000420131.1 ENST00000414630.2 ENST00000453049.1 ENST00000428819.1 ENST00000448598.1 ENST00000449088.3 ENST00000430405.3 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 | 0.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |