Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
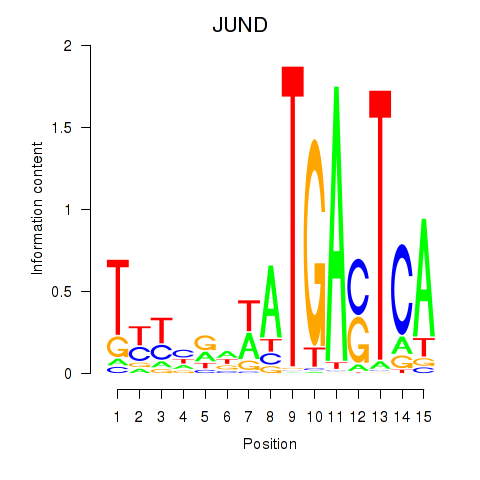
Results for JUND
Z-value: 0.46
Transcription factors associated with JUND
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUND
|
ENSG00000130522.4 | JunD proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUND | hg19_v2_chr19_-_18392422_18392440 | -0.53 | 4.7e-01 | Click! |
Activity profile of JUND motif
Sorted Z-values of JUND motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_52695617 | 0.26 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr12_-_102591604 | 0.21 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr5_-_140013224 | 0.19 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr12_-_112443830 | 0.18 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr1_-_153513170 | 0.17 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr1_+_28527070 | 0.16 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr9_+_34992846 | 0.16 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr15_-_50838888 | 0.14 |
ENST00000532404.1
|
USP50
|
ubiquitin specific peptidase 50 |
| chr12_-_8380183 | 0.14 |
ENST00000442295.2
ENST00000307435.6 ENST00000538603.1 |
FAM90A1
|
family with sequence similarity 90, member A1 |
| chr21_-_43786634 | 0.13 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr14_+_94492674 | 0.11 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr12_+_9142131 | 0.11 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr17_-_25568687 | 0.11 |
ENST00000581944.1
|
RP11-663N22.1
|
RP11-663N22.1 |
| chr8_-_38008783 | 0.11 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr1_+_207262881 | 0.11 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr13_-_30160925 | 0.11 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_-_82359662 | 0.11 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr1_+_223889285 | 0.10 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr19_-_11545920 | 0.10 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr1_+_159796534 | 0.10 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr5_-_140013275 | 0.10 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr4_-_84035905 | 0.09 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr12_-_112443767 | 0.09 |
ENST00000550233.1
ENST00000546962.1 ENST00000550800.2 |
TMEM116
|
transmembrane protein 116 |
| chr17_+_28256874 | 0.09 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr6_+_63921351 | 0.09 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_-_49395705 | 0.08 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr1_+_26605618 | 0.08 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr2_-_145275109 | 0.08 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_4719072 | 0.08 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr16_+_57844549 | 0.08 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr19_-_46285646 | 0.08 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr18_+_9885961 | 0.08 |
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr17_-_73150629 | 0.07 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr17_+_65375082 | 0.07 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_-_23426231 | 0.07 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr22_-_37545972 | 0.07 |
ENST00000216223.5
|
IL2RB
|
interleukin 2 receptor, beta |
| chr11_+_59522837 | 0.07 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr3_-_170587974 | 0.07 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr12_-_74796291 | 0.07 |
ENST00000551726.1
|
RP11-81H3.2
|
RP11-81H3.2 |
| chr19_+_13842559 | 0.07 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr6_-_41673552 | 0.07 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr10_+_127661942 | 0.07 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr5_+_145583156 | 0.07 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr9_-_95432536 | 0.06 |
ENST00000287996.3
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr22_-_36761081 | 0.06 |
ENST00000456729.1
ENST00000401701.1 |
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr17_+_41052808 | 0.06 |
ENST00000592383.1
ENST00000253801.2 ENST00000585489.1 |
G6PC
|
glucose-6-phosphatase, catalytic subunit |
| chr11_+_67250490 | 0.06 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr17_-_70417365 | 0.06 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr1_-_117753540 | 0.06 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr1_-_54637997 | 0.06 |
ENST00000536061.1
|
AL357673.1
|
CDNA: FLJ21031 fis, clone CAE07336; HCG1780521; Uncharacterized protein |
| chr14_+_73525265 | 0.06 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr6_-_32145861 | 0.06 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr16_-_67281413 | 0.06 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr18_+_9885934 | 0.06 |
ENST00000357775.5
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr18_-_74839891 | 0.06 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr15_+_89182178 | 0.05 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_+_85936295 | 0.05 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr4_-_84035868 | 0.05 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr6_+_151358048 | 0.05 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr3_-_98241713 | 0.05 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr16_+_23847339 | 0.05 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr10_-_14996017 | 0.05 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr22_-_43398442 | 0.05 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_-_141061759 | 0.05 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr2_-_152830479 | 0.05 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_-_235116495 | 0.05 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr1_-_223853425 | 0.05 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr3_-_98241760 | 0.04 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr1_-_1342617 | 0.04 |
ENST00000482352.1
ENST00000344843.7 |
MRPL20
|
mitochondrial ribosomal protein L20 |
| chr4_+_77356248 | 0.04 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr15_+_89181974 | 0.04 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr20_+_48429356 | 0.04 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr8_-_141774467 | 0.04 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr6_+_32146131 | 0.04 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr16_+_31366455 | 0.04 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr5_-_141061777 | 0.04 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr15_-_59949693 | 0.04 |
ENST00000396063.1
ENST00000396064.3 ENST00000484743.1 ENST00000559706.1 ENST00000396060.2 |
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr16_-_72206034 | 0.04 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr7_-_33080506 | 0.04 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr16_+_28996364 | 0.04 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr3_-_105588231 | 0.03 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr16_+_15596123 | 0.03 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr2_-_231789941 | 0.03 |
ENST00000392040.1
ENST00000438398.1 |
GPR55
|
G protein-coupled receptor 55 |
| chr1_+_41204506 | 0.03 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chrX_-_154563889 | 0.03 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr3_-_18466026 | 0.03 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr10_-_74114714 | 0.03 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr1_-_205053645 | 0.03 |
ENST00000367167.3
|
TMEM81
|
transmembrane protein 81 |
| chr12_-_95009837 | 0.03 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr17_-_54893250 | 0.03 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr2_-_145275211 | 0.03 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_69811858 | 0.03 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr7_+_100663353 | 0.03 |
ENST00000306151.4
|
MUC17
|
mucin 17, cell surface associated |
| chr6_+_150070857 | 0.03 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_-_154941350 | 0.03 |
ENST00000444179.1
ENST00000414115.1 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr14_-_23426270 | 0.03 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr18_+_43319467 | 0.03 |
ENST00000591541.1
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr1_+_227751231 | 0.03 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr5_-_60140009 | 0.03 |
ENST00000505959.1
|
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr10_-_14996321 | 0.03 |
ENST00000378289.4
|
DCLRE1C
|
DNA cross-link repair 1C |
| chr1_-_223853348 | 0.03 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr4_+_95128996 | 0.02 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr20_-_36794902 | 0.02 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr14_-_23426337 | 0.02 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr11_-_66103867 | 0.02 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_+_7590734 | 0.02 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr12_+_62654155 | 0.02 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr5_-_96518907 | 0.02 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr17_-_45899126 | 0.02 |
ENST00000007414.3
ENST00000392507.3 |
OSBPL7
|
oxysterol binding protein-like 7 |
| chr3_-_122512619 | 0.02 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chrX_+_133371077 | 0.02 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr3_+_158991025 | 0.02 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chrY_+_15016725 | 0.02 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr16_+_19467772 | 0.02 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr12_-_53343602 | 0.02 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr17_-_59668550 | 0.02 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr6_+_159290917 | 0.02 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr4_-_123542224 | 0.02 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr8_+_85097110 | 0.02 |
ENST00000517638.1
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein-like |
| chr18_+_55888767 | 0.02 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_-_46285736 | 0.02 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chr1_-_11918988 | 0.02 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr20_+_48429233 | 0.02 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr14_+_58765103 | 0.02 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr10_-_14996070 | 0.02 |
ENST00000378258.1
ENST00000453695.2 ENST00000378246.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chrX_+_154254960 | 0.02 |
ENST00000369498.3
|
FUNDC2
|
FUN14 domain containing 2 |
| chr3_-_11762202 | 0.02 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr3_+_127770455 | 0.02 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr5_-_138533973 | 0.02 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr10_+_18549645 | 0.01 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr11_-_2162162 | 0.01 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr5_-_41794313 | 0.01 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr2_+_161993465 | 0.01 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr20_+_30946106 | 0.01 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr3_+_111758553 | 0.01 |
ENST00000419127.1
|
TMPRSS7
|
transmembrane protease, serine 7 |
| chr10_+_69865866 | 0.01 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr5_-_60140089 | 0.01 |
ENST00000507047.1
ENST00000438340.1 ENST00000425382.1 ENST00000508821.1 |
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr1_+_236687881 | 0.01 |
ENST00000526634.1
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr12_-_53343633 | 0.01 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr3_-_46608010 | 0.01 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr3_-_37216055 | 0.01 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr14_+_58765305 | 0.01 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr11_-_18343669 | 0.01 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr18_+_32290218 | 0.01 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr14_-_23426322 | 0.01 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr11_-_18343725 | 0.01 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr16_+_28996114 | 0.01 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr1_+_15256230 | 0.01 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr10_-_30024716 | 0.01 |
ENST00000375398.2
ENST00000375400.3 |
SVIL
|
supervillin |
| chr4_-_104119528 | 0.01 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr11_-_66103932 | 0.01 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr8_-_17752912 | 0.01 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr11_-_6008215 | 0.01 |
ENST00000332249.4
|
OR52L1
|
olfactory receptor, family 52, subfamily L, member 1 |
| chr15_-_42565606 | 0.01 |
ENST00000307216.6
ENST00000448392.1 |
TMEM87A
|
transmembrane protein 87A |
| chr6_+_32146268 | 0.01 |
ENST00000427134.2
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr8_+_128426535 | 0.01 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr10_+_17270214 | 0.01 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr8_-_17752996 | 0.01 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr22_-_39715600 | 0.01 |
ENST00000427905.1
ENST00000402527.1 ENST00000216146.4 |
RPL3
|
ribosomal protein L3 |
| chr3_+_184032283 | 0.01 |
ENST00000346169.2
ENST00000414031.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chrX_+_51942963 | 0.01 |
ENST00000375625.3
|
RP11-363G10.2
|
RP11-363G10.2 |
| chr12_-_76817036 | 0.01 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr2_+_181988560 | 0.01 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr12_+_54891495 | 0.01 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr15_-_74504597 | 0.01 |
ENST00000416286.3
|
STRA6
|
stimulated by retinoic acid 6 |
| chr11_-_14913190 | 0.00 |
ENST00000532378.1
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chrX_-_153718953 | 0.00 |
ENST00000369649.4
ENST00000393586.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr18_-_19283649 | 0.00 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr11_-_2162468 | 0.00 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_155860751 | 0.00 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr3_+_169539710 | 0.00 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr17_-_7590745 | 0.00 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr10_+_5005598 | 0.00 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr3_+_184032313 | 0.00 |
ENST00000392537.2
ENST00000444134.1 ENST00000450424.1 ENST00000421110.1 ENST00000382330.3 ENST00000426123.1 ENST00000350481.5 ENST00000455679.1 ENST00000440448.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr5_+_38148582 | 0.00 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr12_+_7023491 | 0.00 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr12_-_122710514 | 0.00 |
ENST00000540535.1
ENST00000541656.1 |
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr16_+_31366536 | 0.00 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr20_-_36794938 | 0.00 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr15_-_74504560 | 0.00 |
ENST00000449139.2
|
STRA6
|
stimulated by retinoic acid 6 |
| chr12_+_12938541 | 0.00 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_+_160709076 | 0.00 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr19_+_11546093 | 0.00 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr18_+_9885760 | 0.00 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
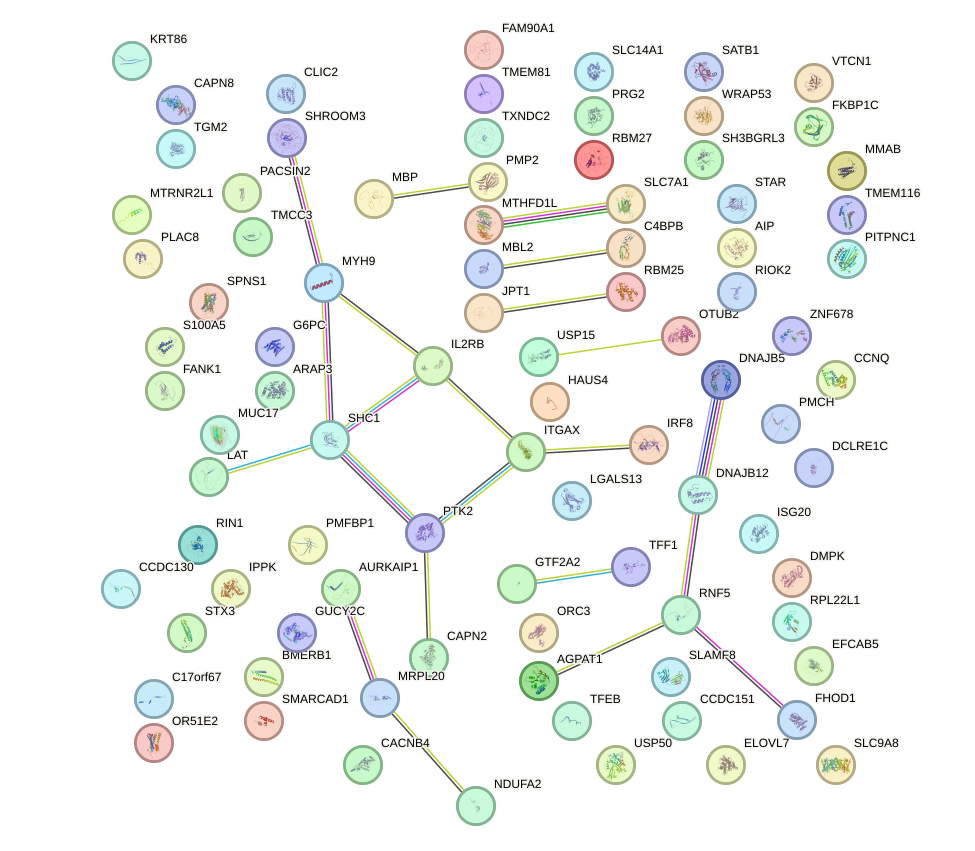
Network of associatons between targets according to the STRING database.
First level regulatory network of JUND
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.1 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0097513 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |