Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
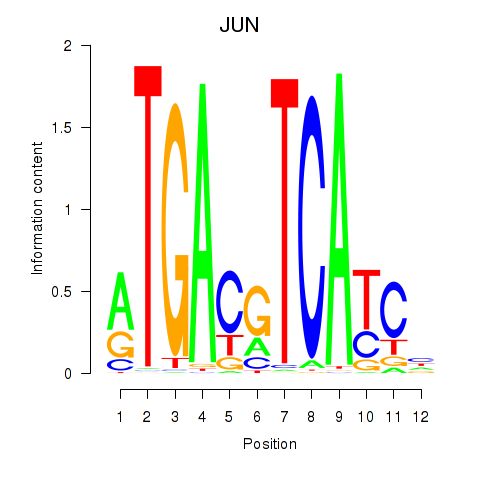
Results for JUN
Z-value: 0.79
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.5 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg19_v2_chr1_-_59249732_59249785 | 0.57 | 4.3e-01 | Click! |
Activity profile of JUN motif
Sorted Z-values of JUN motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_27782788 | 0.79 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr14_-_24732738 | 0.63 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr11_-_9482010 | 0.62 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr22_-_44258360 | 0.62 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr19_+_49109990 | 0.60 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr17_-_43210580 | 0.53 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr6_-_27782548 | 0.51 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr6_+_30749649 | 0.51 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr16_+_3068393 | 0.47 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr6_-_26285737 | 0.47 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr19_-_51014460 | 0.40 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr3_-_134092561 | 0.39 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr16_+_2564254 | 0.39 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr19_+_55996316 | 0.37 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr19_+_47616682 | 0.36 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr19_-_51014588 | 0.35 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr19_-_51014345 | 0.35 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr13_+_113030658 | 0.35 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr1_-_153917700 | 0.33 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr2_-_220408260 | 0.33 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr6_+_26204825 | 0.33 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr22_+_38597889 | 0.33 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr19_-_50979981 | 0.32 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr19_-_47616992 | 0.32 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr5_+_139055021 | 0.31 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr10_-_90967063 | 0.31 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr16_+_29911666 | 0.30 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr11_+_122709200 | 0.29 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr19_+_57791419 | 0.29 |
ENST00000537645.1
|
ZNF460
|
zinc finger protein 460 |
| chr1_+_36023370 | 0.29 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr11_+_66610883 | 0.29 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr8_-_21966893 | 0.29 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr8_-_103876340 | 0.29 |
ENST00000518353.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr14_+_74003818 | 0.28 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr17_+_73089382 | 0.28 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr5_-_139930713 | 0.28 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr11_+_29181503 | 0.28 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr19_+_50979753 | 0.27 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr2_-_220408430 | 0.27 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr11_+_66059339 | 0.26 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chrX_+_48398053 | 0.26 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr16_+_29911864 | 0.25 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr8_-_18711866 | 0.25 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr19_+_46009837 | 0.25 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr6_+_151358048 | 0.25 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr7_+_100551239 | 0.25 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr16_+_30662050 | 0.24 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr1_+_36024107 | 0.23 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr11_+_392587 | 0.23 |
ENST00000534401.1
|
PKP3
|
plakophilin 3 |
| chr20_-_44600810 | 0.23 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr19_+_49122548 | 0.23 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr5_-_56778635 | 0.23 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr3_-_107941209 | 0.22 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr9_-_130477912 | 0.22 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chrX_+_152240819 | 0.22 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr1_-_204121102 | 0.22 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr11_+_818902 | 0.22 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chr16_+_30662085 | 0.22 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr15_+_75287861 | 0.21 |
ENST00000425597.3
ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5
|
secretory carrier membrane protein 5 |
| chr17_-_40828969 | 0.21 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr19_+_44100632 | 0.21 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr1_-_45140074 | 0.21 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr2_-_207082748 | 0.21 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr6_-_34664612 | 0.20 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr19_+_41094612 | 0.20 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr20_+_3190006 | 0.20 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr19_+_42772659 | 0.20 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr6_+_42123141 | 0.20 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr14_-_77495007 | 0.20 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr14_-_55738788 | 0.20 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr12_-_52761262 | 0.20 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr19_+_24009879 | 0.20 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr16_+_2563871 | 0.20 |
ENST00000330398.4
ENST00000568562.1 ENST00000569317.1 |
ATP6V0C
ATP6C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr6_-_31940065 | 0.19 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr5_-_139943830 | 0.19 |
ENST00000412920.3
ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr11_-_117699413 | 0.19 |
ENST00000528014.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr7_+_99905325 | 0.19 |
ENST00000332397.6
ENST00000437326.2 |
SPDYE3
|
speedy/RINGO cell cycle regulator family member E3 |
| chr12_-_100660833 | 0.19 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr1_-_204121298 | 0.19 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr10_+_134210672 | 0.19 |
ENST00000305233.5
ENST00000368609.4 |
PWWP2B
|
PWWP domain containing 2B |
| chr17_+_685513 | 0.19 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr12_+_43086018 | 0.19 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr17_-_43209862 | 0.18 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr11_+_6226782 | 0.18 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr19_+_10362882 | 0.18 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr5_+_174151536 | 0.18 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr16_-_4588762 | 0.18 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr11_+_45825616 | 0.18 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr17_+_27071002 | 0.18 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr6_+_118869452 | 0.18 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr6_+_28048753 | 0.18 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr1_+_153750622 | 0.18 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr22_+_51176624 | 0.17 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr2_-_224467002 | 0.17 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr10_+_35484793 | 0.17 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr19_+_36024310 | 0.17 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chrX_-_152245978 | 0.17 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr19_-_40919271 | 0.17 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr5_+_139055055 | 0.17 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr3_-_119182453 | 0.17 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr6_+_30029008 | 0.17 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr14_+_105219437 | 0.17 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr17_+_41476327 | 0.17 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr1_+_198608146 | 0.17 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_129592471 | 0.17 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr2_-_98280383 | 0.16 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr8_+_86121448 | 0.16 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr19_-_46088068 | 0.16 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr11_-_62457371 | 0.16 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr5_-_172198190 | 0.16 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr11_+_809647 | 0.16 |
ENST00000321153.4
|
RPLP2
|
ribosomal protein, large, P2 |
| chr16_+_30751953 | 0.16 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr21_+_29911640 | 0.16 |
ENST00000412526.1
ENST00000455939.1 |
LINC00161
|
long intergenic non-protein coding RNA 161 |
| chr11_-_62389621 | 0.16 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr12_-_6982442 | 0.15 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr16_-_5083917 | 0.15 |
ENST00000312251.3
ENST00000381955.3 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr22_-_32026810 | 0.15 |
ENST00000266095.5
ENST00000397500.1 |
PISD
|
phosphatidylserine decarboxylase |
| chr1_-_204121013 | 0.15 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr19_+_57791806 | 0.15 |
ENST00000360338.3
|
ZNF460
|
zinc finger protein 460 |
| chr8_-_30013748 | 0.15 |
ENST00000607315.1
|
RP11-51J9.5
|
RP11-51J9.5 |
| chr3_-_196295385 | 0.15 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr7_+_100273736 | 0.15 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr17_+_38497640 | 0.15 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr17_+_7155819 | 0.15 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr8_-_145754428 | 0.14 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr20_-_3154162 | 0.14 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr5_+_137673200 | 0.14 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr8_-_42065075 | 0.14 |
ENST00000429089.2
ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT
|
plasminogen activator, tissue |
| chr12_+_52668394 | 0.14 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr19_-_2944907 | 0.14 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr6_+_41888926 | 0.14 |
ENST00000230340.4
|
BYSL
|
bystin-like |
| chr2_+_38177575 | 0.14 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr7_+_30174574 | 0.14 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr19_-_19030157 | 0.14 |
ENST00000349893.4
ENST00000351079.4 ENST00000600932.1 ENST00000262812.4 |
COPE
|
coatomer protein complex, subunit epsilon |
| chr10_-_61720640 | 0.14 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr14_-_69261310 | 0.14 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_+_36036477 | 0.14 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr17_-_40829026 | 0.14 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr1_-_16482554 | 0.14 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr1_-_32264356 | 0.14 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr22_+_39898391 | 0.14 |
ENST00000434364.1
|
MIEF1
|
mitochondrial elongation factor 1 |
| chrX_-_7895755 | 0.14 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr7_+_22766766 | 0.14 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr6_+_37400974 | 0.14 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr19_-_49540073 | 0.13 |
ENST00000301407.7
ENST00000601167.1 ENST00000604577.1 ENST00000591656.1 |
CGB1
CTB-60B18.6
|
chorionic gonadotropin, beta polypeptide 1 Choriogonadotropin subunit beta variant 1; Uncharacterized protein |
| chr2_+_74781828 | 0.13 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr17_-_71308119 | 0.13 |
ENST00000439510.2
ENST00000581014.1 ENST00000579611.1 |
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr7_-_45026419 | 0.13 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chrX_+_12993336 | 0.13 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr5_-_175964366 | 0.13 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr20_-_31124186 | 0.13 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr9_-_140115775 | 0.13 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr19_-_35264089 | 0.13 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr12_+_56521840 | 0.13 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr19_+_36119975 | 0.13 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr7_+_143078379 | 0.13 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr2_+_70142232 | 0.13 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr19_-_55574538 | 0.12 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr7_+_4721885 | 0.12 |
ENST00000328914.4
|
FOXK1
|
forkhead box K1 |
| chr15_-_72766533 | 0.12 |
ENST00000562573.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr2_+_241375069 | 0.12 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr2_-_220264703 | 0.12 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr6_-_41888843 | 0.12 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr16_+_2039946 | 0.12 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr7_+_5632436 | 0.12 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chrX_+_12993202 | 0.12 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr1_-_32110467 | 0.12 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr11_+_64107663 | 0.12 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr1_-_1284730 | 0.12 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr19_-_55919087 | 0.12 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr1_+_173837488 | 0.12 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr9_+_97488939 | 0.11 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr12_+_13061894 | 0.11 |
ENST00000540125.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr3_-_50329990 | 0.11 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr19_+_36036583 | 0.11 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr19_+_17830051 | 0.11 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chrX_-_48755030 | 0.11 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr14_+_77228532 | 0.11 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr14_-_23446900 | 0.11 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr19_+_44100727 | 0.11 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr16_-_90038866 | 0.11 |
ENST00000314994.3
|
CENPBD1
|
CENPB DNA-binding domains containing 1 |
| chr17_-_7155775 | 0.11 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr15_-_100273544 | 0.11 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr17_-_72864739 | 0.11 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr6_-_10694766 | 0.10 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr2_+_241544834 | 0.10 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr9_+_131903916 | 0.10 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr3_+_184081137 | 0.10 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_+_93651296 | 0.10 |
ENST00000283534.4
ENST00000557574.1 |
TMEM251
RP11-371E8.4
|
transmembrane protein 251 Uncharacterized protein |
| chrX_-_153602991 | 0.10 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr3_-_47934234 | 0.10 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr12_-_52887034 | 0.10 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr5_+_139944396 | 0.10 |
ENST00000514199.1
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr19_-_45735138 | 0.10 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr12_+_56522001 | 0.10 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr11_+_3819049 | 0.10 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr5_+_32174483 | 0.10 |
ENST00000606994.1
|
CTD-2186M15.3
|
CTD-2186M15.3 |
| chr8_-_10697370 | 0.10 |
ENST00000314787.3
ENST00000426190.2 ENST00000519088.1 |
PINX1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr3_+_187871659 | 0.10 |
ENST00000416784.1
ENST00000430340.1 ENST00000414139.1 ENST00000454789.1 |
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr3_-_9994021 | 0.10 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of JUN
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.3 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.2 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.2 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.3 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0090208 | positive regulation of triglyceride biosynthetic process(GO:0010867) positive regulation of triglyceride metabolic process(GO:0090208) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |