Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
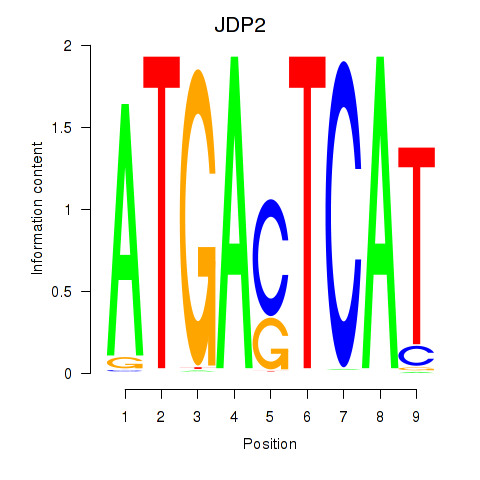
Results for JDP2
Z-value: 0.35
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JDP2 | hg19_v2_chr14_+_75894714_75894749 | -0.96 | 4.4e-02 | Click! |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_75083313 | 0.25 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr11_+_28724129 | 0.22 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr1_-_12679171 | 0.20 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr16_+_3692931 | 0.18 |
ENST00000407479.1
|
DNASE1
|
deoxyribonuclease I |
| chr7_+_30960915 | 0.17 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr8_+_40018977 | 0.17 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr20_+_4667094 | 0.16 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr2_+_208104351 | 0.15 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr12_-_105352047 | 0.14 |
ENST00000432951.1
ENST00000415674.1 ENST00000424946.1 |
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr1_-_160231451 | 0.13 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr8_+_86157699 | 0.13 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr12_-_75784669 | 0.12 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr17_-_42994283 | 0.12 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr16_-_30997533 | 0.12 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr19_-_56047661 | 0.11 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr1_+_213224572 | 0.10 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr17_+_75315654 | 0.10 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr7_-_47492782 | 0.10 |
ENST00000413551.1
|
TNS3
|
tensin 3 |
| chr2_-_121624973 | 0.09 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr14_-_54317532 | 0.09 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr6_-_56819385 | 0.09 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr10_-_75385711 | 0.09 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr11_-_8739383 | 0.09 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr16_-_4164027 | 0.09 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr4_-_178169927 | 0.08 |
ENST00000512516.1
|
RP11-487E13.1
|
Uncharacterized protein |
| chr6_-_161695042 | 0.08 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr3_+_98699880 | 0.08 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr12_+_113860160 | 0.08 |
ENST00000553248.1
ENST00000345635.4 ENST00000547802.1 |
SDSL
|
serine dehydratase-like |
| chr18_-_74207146 | 0.08 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr15_+_57211318 | 0.08 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chrX_-_154493791 | 0.08 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr10_-_90611566 | 0.08 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr2_-_65593784 | 0.08 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr14_-_51562037 | 0.07 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr2_+_7073174 | 0.07 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr19_-_51872233 | 0.07 |
ENST00000601435.1
ENST00000291715.1 |
CLDND2
|
claudin domain containing 2 |
| chr17_-_55162360 | 0.07 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr3_+_57094469 | 0.07 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr2_+_47596634 | 0.07 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chr2_+_27505260 | 0.07 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr17_+_30771279 | 0.07 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr2_+_87754989 | 0.07 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr14_-_65769392 | 0.07 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr12_-_105352080 | 0.07 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr12_+_113860042 | 0.06 |
ENST00000403593.4
|
SDSL
|
serine dehydratase-like |
| chr10_-_116418053 | 0.06 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr16_-_31161380 | 0.06 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr4_+_170581213 | 0.06 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr16_+_57662596 | 0.06 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_-_19739321 | 0.06 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr9_+_118950325 | 0.06 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr12_-_15104040 | 0.06 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr20_+_42136308 | 0.06 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr20_+_43029911 | 0.06 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr2_+_233562015 | 0.06 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr2_+_152214098 | 0.06 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr4_-_122085469 | 0.06 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr10_-_21463116 | 0.06 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr4_-_987217 | 0.06 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_-_90610200 | 0.05 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr8_+_22435762 | 0.05 |
ENST00000456545.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr20_-_17539456 | 0.05 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr17_+_25799008 | 0.05 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr13_-_41593425 | 0.05 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr17_-_1508379 | 0.05 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr19_+_38880695 | 0.05 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr7_-_135433460 | 0.05 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr9_+_44867571 | 0.05 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr2_-_175711978 | 0.05 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr3_+_138153451 | 0.05 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr11_-_57417367 | 0.05 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr1_-_93257951 | 0.05 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr3_+_183903811 | 0.05 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr18_+_61442629 | 0.04 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr4_+_76481258 | 0.04 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr9_-_112179990 | 0.04 |
ENST00000394827.3
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr12_-_719573 | 0.04 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr3_-_195997410 | 0.04 |
ENST00000419333.1
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr2_-_11605966 | 0.04 |
ENST00000307236.4
ENST00000542100.1 ENST00000546212.1 |
E2F6
|
E2F transcription factor 6 |
| chr1_-_153521597 | 0.04 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_+_75446629 | 0.04 |
ENST00000588958.2
ENST00000586128.1 |
SEPT9
|
septin 9 |
| chr7_+_55177416 | 0.04 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr16_+_28996416 | 0.04 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr12_-_53297432 | 0.04 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr8_-_27472198 | 0.04 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr5_+_145317356 | 0.04 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr6_+_56819773 | 0.04 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr7_+_102191679 | 0.04 |
ENST00000507918.1
ENST00000432940.1 |
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chr11_-_82708519 | 0.04 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr8_-_67974552 | 0.04 |
ENST00000357849.4
|
COPS5
|
COP9 signalosome subunit 5 |
| chr2_-_11606275 | 0.04 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr7_+_134528635 | 0.04 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr17_+_75446819 | 0.04 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
| chr16_+_57662527 | 0.04 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_-_31288398 | 0.04 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr10_+_1102303 | 0.03 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr7_-_80551671 | 0.03 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr4_+_159727272 | 0.03 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr16_+_14280564 | 0.03 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr3_+_124223586 | 0.03 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr17_+_47287749 | 0.03 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr1_+_154378049 | 0.03 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr12_+_10365404 | 0.03 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr19_-_35992780 | 0.03 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr8_-_123139423 | 0.03 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr7_+_1126437 | 0.03 |
ENST00000413368.1
ENST00000397092.1 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr1_-_244006528 | 0.03 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_150721570 | 0.03 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr5_+_145583107 | 0.03 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr1_-_108231101 | 0.03 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr16_+_28996114 | 0.03 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr7_+_120590803 | 0.03 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr11_-_82708435 | 0.03 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr14_+_32476072 | 0.03 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chrX_-_15288154 | 0.03 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr5_-_75919253 | 0.03 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr12_+_10365082 | 0.03 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr3_+_191046810 | 0.03 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr5_-_149492904 | 0.03 |
ENST00000286301.3
ENST00000511344.1 |
CSF1R
|
colony stimulating factor 1 receptor |
| chr1_-_151965048 | 0.03 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr4_-_36246060 | 0.03 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_-_175712270 | 0.03 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr4_+_118349545 | 0.03 |
ENST00000422145.3
ENST00000437514.1 |
AC092661.1
|
AC092661.1 |
| chr17_-_28257080 | 0.03 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr4_+_3388057 | 0.03 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr5_+_177540444 | 0.02 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr10_+_94594351 | 0.02 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr9_+_123906331 | 0.02 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr6_+_10521574 | 0.02 |
ENST00000495262.1
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr11_-_82782861 | 0.02 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr18_+_74207477 | 0.02 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr3_-_48632593 | 0.02 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr2_+_208104497 | 0.02 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chrX_+_41193407 | 0.02 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr7_+_872107 | 0.02 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr7_+_102290772 | 0.02 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr2_+_47596287 | 0.02 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chrX_+_133371077 | 0.02 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr13_-_67802549 | 0.02 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr14_+_73525229 | 0.02 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr1_+_156096336 | 0.02 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr3_+_48507621 | 0.02 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr1_+_154377669 | 0.02 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr17_+_75315534 | 0.02 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr5_+_72143988 | 0.02 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr13_-_36429763 | 0.02 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr4_+_169013666 | 0.02 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr7_-_135433534 | 0.02 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr1_-_205744574 | 0.02 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr5_-_138534071 | 0.02 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr6_+_56820018 | 0.02 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr11_+_44069531 | 0.02 |
ENST00000378832.1
|
ACCSL
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like |
| chr14_+_73525144 | 0.02 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr1_-_45272951 | 0.02 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr5_+_179247759 | 0.02 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr7_+_23286182 | 0.02 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr10_-_120514720 | 0.02 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr15_+_41062159 | 0.01 |
ENST00000344320.6
|
C15orf62
|
chromosome 15 open reading frame 62 |
| chr9_+_127023704 | 0.01 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr2_+_28615669 | 0.01 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr3_+_28283069 | 0.01 |
ENST00000466830.1
ENST00000423894.1 |
CMC1
|
C-x(9)-C motif containing 1 |
| chr2_+_169926047 | 0.01 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr17_-_38956205 | 0.01 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr1_-_45476944 | 0.01 |
ENST00000372172.4
|
HECTD3
|
HECT domain containing E3 ubiquitin protein ligase 3 |
| chr1_+_152881014 | 0.01 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr2_+_10262442 | 0.01 |
ENST00000360566.2
|
RRM2
|
ribonucleotide reductase M2 |
| chr11_-_75201791 | 0.01 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr21_+_42742429 | 0.01 |
ENST00000418103.1
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr17_-_73775839 | 0.01 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr3_+_19988885 | 0.01 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr14_-_38028689 | 0.01 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr13_+_32838801 | 0.01 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr10_+_134901388 | 0.01 |
ENST00000392607.3
|
GPR123
|
G protein-coupled receptor 123 |
| chr2_-_175712479 | 0.01 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr11_+_57308979 | 0.01 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr4_+_159727222 | 0.01 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr2_-_220173685 | 0.01 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr14_-_51561784 | 0.01 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr11_+_82783097 | 0.01 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr4_-_146019693 | 0.01 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr17_+_48712206 | 0.01 |
ENST00000427699.1
ENST00000285238.8 |
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr1_-_153521714 | 0.01 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_-_122342966 | 0.01 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr6_+_292051 | 0.01 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr18_+_2846972 | 0.01 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chrX_+_153770421 | 0.01 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr11_+_58874701 | 0.01 |
ENST00000529618.1
ENST00000534403.1 ENST00000343597.3 |
FAM111B
|
family with sequence similarity 111, member B |
| chr3_-_48601206 | 0.01 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr9_-_21335356 | 0.01 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr6_-_31745037 | 0.01 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr1_+_198189921 | 0.01 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr3_-_151034734 | 0.01 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_73109339 | 0.01 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr14_-_74226961 | 0.01 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr10_-_4285923 | 0.01 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr4_+_120056939 | 0.01 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr3_-_114343768 | 0.01 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_149806105 | 0.01 |
ENST00000389942.5
ENST00000416573.2 ENST00000542614.1 ENST00000409806.3 |
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr11_+_101983176 | 0.00 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr4_+_86525299 | 0.00 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071288 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.0 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |