Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
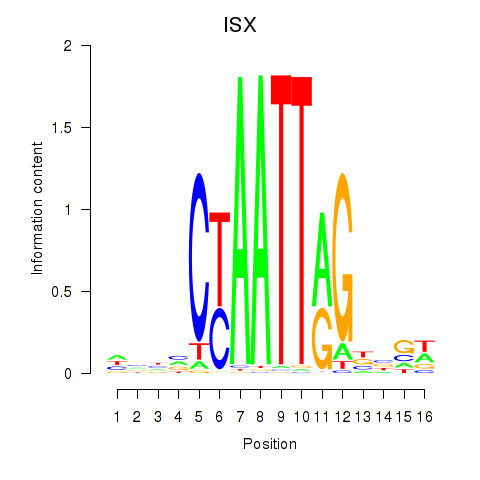
Results for ISX
Z-value: 0.29
Transcription factors associated with ISX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISX
|
ENSG00000175329.8 | intestine specific homeobox |
Activity profile of ISX motif
Sorted Z-values of ISX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_75199213 | 0.13 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr9_-_75488984 | 0.13 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr16_+_12059091 | 0.11 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr7_-_86849836 | 0.10 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr17_-_18266818 | 0.09 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr1_+_32666188 | 0.09 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr19_-_4717835 | 0.08 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr22_+_46476192 | 0.08 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr14_-_88200641 | 0.07 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr20_-_50418947 | 0.07 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr11_-_95523500 | 0.07 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr8_+_110552046 | 0.07 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr12_-_10978957 | 0.07 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr16_+_3333443 | 0.06 |
ENST00000572748.1
ENST00000573578.1 ENST00000574253.1 |
ZNF263
|
zinc finger protein 263 |
| chr3_-_129147432 | 0.06 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr14_-_51027838 | 0.06 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_120839005 | 0.06 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr14_+_70233810 | 0.05 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chrX_-_47509994 | 0.05 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr4_+_66536248 | 0.05 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr1_+_206138457 | 0.05 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr1_-_143913143 | 0.05 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr9_+_130159433 | 0.05 |
ENST00000451404.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr8_+_38831683 | 0.05 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr17_-_12920994 | 0.05 |
ENST00000609101.1
|
ELAC2
|
elaC ribonuclease Z 2 |
| chr17_-_12920907 | 0.05 |
ENST00000609757.1
ENST00000581499.2 ENST00000580504.1 |
ELAC2
|
elaC ribonuclease Z 2 |
| chr3_-_15563229 | 0.05 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr11_+_77532233 | 0.05 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr1_-_158173659 | 0.05 |
ENST00000415019.1
|
RP11-404O13.5
|
RP11-404O13.5 |
| chr6_+_3259148 | 0.05 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr3_-_186288097 | 0.05 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr4_-_48018680 | 0.05 |
ENST00000513178.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr5_-_98262240 | 0.05 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr1_+_28261621 | 0.05 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr2_-_183387064 | 0.04 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_+_91461413 | 0.04 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr7_-_112635675 | 0.04 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr14_-_24711764 | 0.04 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_+_5712234 | 0.04 |
ENST00000414641.1
|
TRIM22
|
tripartite motif containing 22 |
| chr17_-_7307358 | 0.04 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr16_-_3493528 | 0.04 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr17_-_38821373 | 0.04 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr2_+_177015122 | 0.04 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr1_-_36916066 | 0.04 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr12_+_104680793 | 0.04 |
ENST00000529546.1
ENST00000529751.1 ENST00000540716.1 ENST00000528079.2 ENST00000526580.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr16_+_3070356 | 0.04 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr20_-_50418972 | 0.04 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr6_-_30080876 | 0.04 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr1_+_87595433 | 0.04 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr6_-_32095968 | 0.04 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr12_-_10022735 | 0.04 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr22_+_24309089 | 0.04 |
ENST00000215770.5
|
DDTL
|
D-dopachrome tautomerase-like |
| chr22_+_21400229 | 0.04 |
ENST00000342608.4
ENST00000543388.1 ENST00000442047.1 |
AC002472.13
|
Leucine-rich repeat-containing protein LOC400891 |
| chr5_-_137374288 | 0.04 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr17_+_46970134 | 0.03 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr14_-_73925225 | 0.03 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr3_-_113775328 | 0.03 |
ENST00000483766.1
ENST00000545063.1 ENST00000491000.1 ENST00000295878.3 |
KIAA1407
|
KIAA1407 |
| chr12_+_16500571 | 0.03 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr8_+_9953061 | 0.03 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr21_+_35014829 | 0.03 |
ENST00000451686.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr21_+_35014706 | 0.03 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr21_+_35014783 | 0.03 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr5_-_177580855 | 0.03 |
ENST00000514354.1
ENST00000511078.1 |
NHP2
|
NHP2 ribonucleoprotein |
| chr17_+_50939459 | 0.03 |
ENST00000412360.1
|
AC102948.2
|
Uncharacterized protein |
| chr6_-_111927062 | 0.03 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr14_+_104182061 | 0.03 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr1_-_21620877 | 0.03 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_-_23670752 | 0.03 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr17_+_46970127 | 0.03 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_+_7014126 | 0.03 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr6_-_30080863 | 0.03 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr1_-_161208013 | 0.03 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_+_150929550 | 0.03 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr6_+_64345698 | 0.03 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr12_+_70574088 | 0.03 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chrX_-_47509887 | 0.03 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chrX_-_153363125 | 0.03 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr2_-_228244013 | 0.03 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr2_+_242089833 | 0.03 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr5_-_177580933 | 0.03 |
ENST00000274606.3
|
NHP2
|
NHP2 ribonucleoprotein |
| chr19_-_51071302 | 0.03 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr19_-_3557570 | 0.03 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr12_+_104680659 | 0.03 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr1_-_197115818 | 0.02 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr6_+_127898312 | 0.02 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr9_+_104296163 | 0.02 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr4_-_70080449 | 0.02 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr6_-_31125850 | 0.02 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr11_-_62521614 | 0.02 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chrY_+_14813160 | 0.02 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr14_-_24711865 | 0.02 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr20_+_24449821 | 0.02 |
ENST00000376862.3
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr8_+_9953214 | 0.02 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr6_-_138833630 | 0.02 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chrX_-_153363188 | 0.02 |
ENST00000303391.6
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr9_+_99212403 | 0.02 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr1_-_150849208 | 0.02 |
ENST00000358595.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr12_-_120966943 | 0.02 |
ENST00000552443.1
ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5
|
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr6_+_34204642 | 0.02 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_-_112281875 | 0.02 |
ENST00000527621.1
ENST00000534365.1 ENST00000357260.5 |
FAM212B
|
family with sequence similarity 212, member B |
| chr14_-_94789663 | 0.02 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr17_-_43045439 | 0.02 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr13_+_40229764 | 0.02 |
ENST00000416691.1
ENST00000422759.2 ENST00000455146.3 |
COG6
|
component of oligomeric golgi complex 6 |
| chr3_-_151034734 | 0.02 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr1_-_23670813 | 0.02 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chrX_+_43515467 | 0.02 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr17_-_79650818 | 0.02 |
ENST00000397498.4
|
ARL16
|
ADP-ribosylation factor-like 16 |
| chr4_-_171012844 | 0.02 |
ENST00000502392.1
|
AADAT
|
aminoadipate aminotransferase |
| chr14_-_73924954 | 0.02 |
ENST00000555307.1
ENST00000554818.1 |
NUMB
|
numb homolog (Drosophila) |
| chr8_+_22132847 | 0.02 |
ENST00000521356.1
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr6_-_85474219 | 0.02 |
ENST00000369663.5
|
TBX18
|
T-box 18 |
| chr4_-_174255536 | 0.02 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr10_+_114206956 | 0.02 |
ENST00000432306.1
ENST00000393077.2 |
VTI1A
|
vesicle transport through interaction with t-SNAREs 1A |
| chr7_+_148982396 | 0.02 |
ENST00000418158.2
|
ZNF783
|
zinc finger family member 783 |
| chr5_+_150226085 | 0.02 |
ENST00000522154.1
|
IRGM
|
immunity-related GTPase family, M |
| chr19_+_42041860 | 0.02 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr10_+_124914285 | 0.01 |
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr8_+_22132810 | 0.01 |
ENST00000356766.6
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr10_-_104001231 | 0.01 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr6_+_148663729 | 0.01 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr17_+_40440481 | 0.01 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr1_-_159832438 | 0.01 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr5_+_140557371 | 0.01 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr1_+_162336686 | 0.01 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr14_+_104182105 | 0.01 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr2_+_87135076 | 0.01 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr1_-_150849047 | 0.01 |
ENST00000354396.2
ENST00000505755.1 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr2_+_135809835 | 0.01 |
ENST00000264158.8
ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr15_-_55700522 | 0.01 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr14_-_53258314 | 0.01 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr8_-_7243080 | 0.01 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr5_+_125935960 | 0.01 |
ENST00000297540.4
|
PHAX
|
phosphorylated adaptor for RNA export |
| chrX_+_19362011 | 0.01 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr1_-_68698222 | 0.01 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr22_+_36044411 | 0.01 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr1_-_23670817 | 0.01 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr8_+_21881636 | 0.01 |
ENST00000520125.1
ENST00000521157.1 ENST00000397940.1 ENST00000522813.1 |
NPM2
|
nucleophosmin/nucleoplasmin 2 |
| chr11_-_108093329 | 0.01 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr14_-_104181771 | 0.01 |
ENST00000554913.1
ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr1_-_150849174 | 0.01 |
ENST00000515192.1
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr14_-_24711470 | 0.01 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr9_+_104296243 | 0.01 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr8_+_9413410 | 0.01 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr1_+_62902308 | 0.01 |
ENST00000339950.4
|
USP1
|
ubiquitin specific peptidase 1 |
| chr9_+_136501478 | 0.01 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr16_+_3070313 | 0.01 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr11_+_57480046 | 0.01 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr2_-_183387430 | 0.01 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_159846399 | 0.01 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr14_+_57671888 | 0.01 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr14_+_39944025 | 0.01 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_+_3000037 | 0.01 |
ENST00000544943.1
ENST00000448120.2 |
TULP3
|
tubby like protein 3 |
| chr8_-_12612962 | 0.01 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr22_+_17956618 | 0.01 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr17_+_59489112 | 0.01 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr14_+_32798462 | 0.01 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_-_66536196 | 0.01 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr17_+_42785976 | 0.01 |
ENST00000393547.2
ENST00000398338.3 |
DBF4B
|
DBF4 homolog B (S. cerevisiae) |
| chr15_-_42500351 | 0.01 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr19_+_50270219 | 0.01 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr15_-_55700457 | 0.01 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr3_+_37284668 | 0.01 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr12_-_50790267 | 0.01 |
ENST00000327337.5
ENST00000543111.1 |
FAM186A
|
family with sequence similarity 186, member A |
| chr17_-_18266765 | 0.01 |
ENST00000354098.3
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr12_+_122688090 | 0.01 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr18_-_67624160 | 0.01 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr10_-_4285923 | 0.01 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr9_+_104296122 | 0.01 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr8_+_110551925 | 0.01 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr19_-_6424783 | 0.01 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr14_-_20801427 | 0.01 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr9_+_34646624 | 0.01 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr10_-_56561022 | 0.01 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr6_+_36164487 | 0.01 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr12_-_51422017 | 0.01 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr6_-_111927449 | 0.01 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr17_-_77005813 | 0.01 |
ENST00000590370.1
ENST00000591625.1 |
CANT1
|
calcium activated nucleotidase 1 |
| chr19_+_36346734 | 0.01 |
ENST00000586102.3
|
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr6_+_28317685 | 0.01 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr17_+_73452545 | 0.01 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr4_-_122686261 | 0.01 |
ENST00000337677.5
|
TMEM155
|
transmembrane protein 155 |
| chr3_-_160823040 | 0.01 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr5_-_95297534 | 0.01 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr6_+_31126291 | 0.01 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr11_-_63381925 | 0.01 |
ENST00000415826.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr3_+_111393501 | 0.00 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_+_177631523 | 0.00 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr4_+_87515454 | 0.00 |
ENST00000427191.2
ENST00000436978.1 ENST00000502971.1 |
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr5_+_177631497 | 0.00 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr18_-_67623906 | 0.00 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr3_-_149095652 | 0.00 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr11_-_77532050 | 0.00 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr12_-_53730147 | 0.00 |
ENST00000536324.2
|
SP7
|
Sp7 transcription factor |
| chr1_+_220267429 | 0.00 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr20_-_35580104 | 0.00 |
ENST00000373694.5
|
SAMHD1
|
SAM domain and HD domain 1 |
| chr2_+_124782857 | 0.00 |
ENST00000431078.1
|
CNTNAP5
|
contactin associated protein-like 5 |
| chr1_+_44115814 | 0.00 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr4_-_186733363 | 0.00 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_33061543 | 0.00 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr20_-_50419055 | 0.00 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |